The attention given to studying GM plants has led to interesting findings pertaining to the detection, persistence and survivability of specific plant genes and proteins in animal systems. Absorption of plant DNA across the intestinal barrier is a natural event as endogenous (native) plant genes have been detected in tissues and products from poultry (Klotz et al. Reference Klotz, Mayer and Einspanier2002; Tony et al. Reference Tony, Butschke, Broll, Grohmann, Zagon, Halle, Danicke, Schauzu, Hafez and Flachowsky2003; Aeschbacher et al. Reference Aeschbacher, Messikommer, Meile and Wenk2005), swine (Reuter & Aulrich, Reference Reuter and Aulrich2003; Nemeth et al. Reference Nemeth, Wurz, Artim, Charlton, Dana, Glenn, Hunst, Jennings, Shilito and Song2004; Mazza et al. Reference Mazza, Soave, Morlacchini, Piva and Marocco2005) and cattle (Einspanier et al. Reference Einspanier, Klotz, Kraft, Aulrich, Poser, Schwagele, Jahreis and Flachowsky2001; Nemeth et al. Reference Nemeth, Wurz, Artim, Charlton, Dana, Glenn, Hunst, Jennings, Shilito and Song2004). The frequency of detection depends on the number of copies of gene ingested and the fragment size of the DNA sequence analysed by PCR. Until recently, however, recombinant plant DNA has not been found in animal tissues. Nemeth et al. (Reference Nemeth, Wurz, Artim, Charlton, Dana, Glenn, Hunst, Jennings, Shilito and Song2004) detected a short fragment (123 bp) of transgenic DNA in swine tissue and our laboratory has detected transgenic DNA (278 bp) in the liver and kidney of swine (Sharma et al. Reference Sharma, Damgaard, Alexander, Dugan, Aalhus, Stanford and McAllister2006). In both studies, positive detection was a rare event. Recently, significantly larger fragments of endogenous (533 bp) and transgenic (519 bp) DNA have been reported with greater frequency in swine tissues (Mazza et al. Reference Mazza, Soave, Morlacchini, Piva and Marocco2005). In each of the studies reporting positive transgene detection, the appearance of transgenic DNA in animal tissues was not statistically more frequent than endogenous DNA.
The range of fragment sizes and discrepancies in occurrence of transgene detection in these studies warrant further investigation into the fate of transgenic DNA in animals. Because major interest lies in the detection of recombinant DNA in animal tissues and the potential for transformation of bacteria by recombinant DNA, it is logical to investigate the amount of transgenic DNA available for each event. The likely place for fragment absorption is the distal small intestine and proximal large intestine, specifically, via the Peyer's patches (Schubbert et al. Reference Schubbert, Renz, Schmitz and Doerfler1997). A previous study has demonstrated that DNA liberated from plant cells is probably rapidly digested by endogenous and microbial nucleases within the small intestine of sheep (Alexander et al. Reference Alexander, Sharma, Deng, Whetsell, Jennings, Wang, Okine, Damgaard and McAllister2004). Although detection of transgenic DNA has been reported throughout the digestive tract of cattle consuming GM feed, the amount of transgenic DNA reaching the small intestine of ruminants has not been reported. Einspanier et al. (Reference Einspanier, Lutz, Rief, Berezina, Zverlov, Schwarz and Mayer2004) attempted to quantify the amount of transgene in digesta from cattle fed GM maize silage, an objective that was not possible due to the low levels of transgene after ensiling. Netherwood et al. (Reference Netherwood, Martìn-Orúe, O'Donnell, Gockling, Graham, Mather and Gilbert2004) were successful in quantifying the number of transgene copies passing to the small intestine of human ileostomists consuming GM soya, and found that up to 3·7 % of the transgene could survive passage.
Roundup Ready® (RR) rapeseed is rendered tolerant to the glyphosate family of herbicides through expression of transgenic DNA from the CP4 strain of Agrobacterium tumefaciens that encodes 5-enolpyruvylshikimate-3-phosphate synthase (cp4 epsps). Our laboratory has published data (Alexander et al. Reference Alexander, Sharma, Okine, Dixon, Forster, Stanford and McAllister2002; Sharma et al. Reference Sharma, Alexander, John, Forster and McAllister2004) demonstrating similarities between RR rapeseed (event GT73) and the parental line from which it is derived, in their digestion by ruminants, and in the patterns of persistence of endogenous DNA from both lines. The focus of the present study, therefore, was on transgenic DNA only and specifically on quantifying the persistence of transgenic DNA in the rumen, at the proximal duodenum, and the faeces of sheep fed forage-based and concentrate-based in diets containing RR rapeseed meal. A second objective was to determine if transgenic DNA could be detected within microbial DNA or in blood.
Materials and methods
Diets, animals and feeding
Transgene stability was assessed using sheep fed forage-based or concentrate-based pelleted diets, both of which contained 15 % (w/w, as fed) rapeseed meal (Table 1). Each diet was prepared using meal from GM RR rapeseed (event GT73) and the non-GM parental line from which it was derived. The rapeseed meal was provided by Monsanto Company (St Louis, MO, USA), and prepared as described previously (Alexander et al. Reference Alexander, Sharma, Okine, Dixon, Forster, Stanford and McAllister2002). Samples of diets were ground to pass a 1·0 mm screen and dried at 135°C for 3 h (Association of Official Analytical Chemists, 1990) for determination of acid-detergent fibre and neutral-detergent fibre content (Van Soest et al. Reference Van Soest, Robertson and Lewis1991; amylase and sodium sulfate included in the neutral-detergent-fibre procedure).
Table 1 Ingredients of forage-based (F) or concentrate-based (C) diets containing Roundup Ready® (RR) or parental (P) rapeseed meal

NDF, neutral-detergent fibre; ADF, acid-detergent fibre.
* Contained NaCl (93·1 %), Mg (1·25 %), Zn (0·9 %), Mn (0·94 %), Cu (0·13 %), Se (0·003 %), K (1·25 %) and Fe (1·25 %).
† Contained vitamin A (3 mg/g), vitamin D (31·25 μg/g) and vitamin E (6·667 mg/g).
‡ Feed pellet binder.
§ Decoquinate (60 g/kg).
The study was conducted using six mature ruminally and duodenally cannulated Canadian Arcott male sheep, assigned randomly to the forage-based or concentrate-based diets (n 3). The sheep were penned individually, with free access to drinking water, and were fed at 08.00 hours daily. They were cared for in accordance with guidelines set by the Canadian Council on Animal Care (Olfert et al. Reference Olfert, Cross and McWilliam1993). Individual consumption of the pelleted diets was measured for the first 3 d of full adaptation, after which feeding was restricted to 95 % of ad libitum intake.
The study comprised three feeding periods (Fig. 1). In period 1, the sheep were adapted to the pelleted diets containing non-GM rapeseed meal over 14 d. For the next 11 d (period 2), the diets containing RR rapeseed meal were fed. From completion of period 2 onward (period 3), feeding of the non-GM diets was resumed. Each sheep remained in its originally assigned diet group throughout the study.

Fig. 1 Feeding scheme in which sheep were assigned to either concentrate-based (C) or forage-based (F) diets (n 3) containing parental or Roundup Ready® rapeseed meal. Throughout the study, sheep were only fed the initially assigned diet (i.e. C or F). The type of rapeseed meal included in the C and F diets in each period is indicated.
Sample collection
Samples of ruminal fluid (RF), duodenal fluid (DF) and faeces were collected as described by Alexander et al. (Reference Alexander, Sharma, Deng, Whetsell, Jennings, Wang, Okine, Damgaard and McAllister2004). Blood samples (5 ml per sheep) were collected via jugular venepuncture. Except where specified, samples from each of the six sheep were processed individually.
On the last day of period 1 (i.e. before any GM rapeseed meal was fed), whole RF and DF were collected from one sheep from each diet group as a source of DNA for inclusion with the standards in real-time PCR assays, to determine the limit of detection (LOD) and to simulate the in vivo matrices of the samples being assessed.
On days 3, 6 and 9 of period 2, RF and DF samples were collected at 12.00 hours, pooled within diet groups, and duplicate 2 ml subsamples were clarified by centrifugation (13 000 g; 10 min) to enable assessment of DNA isolated from the cell-free supernatant fraction. On days 4 and 7, RF and DF were collected at 12.00 hours and pooled within diet type. Subsamples from the composites were frozen for later extraction of DNA, and the remaining samples were used immediately in batch-culture incubations as described later. On days 1, 5 and 9, blood samples were collected at 13.00 hours for the extraction of DNA from 3 ml subsamples (not pooled). Persistence of transgenic DNA in the gastrointestinal tract was studied in unpooled digesta samples (RF, DF, faeces) collected 1, 4, 7, 10, 13, 17, 21, 25, 29, 33, 37, 43 and 49 h after the last feeding of RR diets (i.e. commencing at 09.00 hours on day 11 and continuing into period 3 (days 12 and 13) when feeding of the non-GM diets had resumed).
Batch-culture incubations of ruminal and duodenal micro-organisms
The possibility of transformation of ruminal or duodenal micro-organisms with transgenic DNA was investigated using an anaerobic batch-culture incubation technique based on that described by Wang et al. (Reference Wang, McAllister, Yanke, Xu, Cheeke and Cheng2000). Serum vials (35 ml capacity) were pre-loaded with 300 mg ground, non-GM diets (forage-based or concentrate-based) as substrate and 15 ml pre-warmed anaerobic buffer (Menke et al. Reference Menke, Raab, Salewski, Steinglass, Fritz and Schneider1979). To begin the incubation, 5 ml whole RF or DF (pooled by forage- or concentrate-based diet) were added to duplicate vials containing the corresponding substrate. The vials were sealed and affixed to a rotary shaker. After 18 h of incubation at 39°C, 5 ml of fermentation liquid were used as inoculum into fresh vials containing buffer and substrate (non-GM diets) only, and vials were incubated again for 18 h at 39°C. This process was repeated for a third 18 h subculture, after which the entire contents of each serum vial were freeze-dried for subsequent assessment for the transgene in extracted DNA. The subcultures were undertaken to ensure digestion of any GM plant material present in the original RF or DF inoculum (assumed to be complete after 54 h; Alexander et al. Reference Alexander, Sharma, Okine, Dixon, Forster, Stanford and McAllister2002), in order to preclude false positives in microbial DNA arising from contamination by recombinant plant DNA.
DNA extraction and quantification
A Wizard® Genomic DNA purification kit (Promega Corporation, Madison, WI, USA) was used for extraction of DNA from blood samples according to the manufacturer's protocol. Isolation of DNA from cell-free RF and DF supernatant fractions was performed as follows: 1 ml of the supernatant fraction was extracted three times with an equal volume of chloroform–isoamyl alcohol (24:1), followed by precipitation in 0·6 volume of isopropyl alcohol, washing with 70 % ethanol, and re-suspension in 50 μl TE buffer (10 mM Tris, 1 mM EDTA; pH 8·0). For extraction of DNA from freeze-dried whole RF, whole DF or faecal material, subsamples of approximately 1 g were ground to a fine powder using a planetary micro mill (Pulverisette-7; Fritsch GmbH, Idar-Oberstein, Germany). In the case of batch cultures, the entire freeze-dried sample was ground in the same manner. The DNA from each powder was extracted using the cetyltrimethylammonium bromide (CTAB) method described by Lipp et al. (Reference Lipp, Brodmann and Pietsch1999), with the following modifications: DNA was extracted from 180 mg of sample, and double volumes of each reagent were used at each step. Subsequently, the samples were incubated for 90 min in the CTAB buffer for lysis and the final DNA was re-suspended in 200 μl TE buffer. This same CTAB extraction method was also used to isolate DNA from RR rapeseed meal for use as positive controls and real-time PCR standards. All DNA was quantified fluorometrically using the Quant-iT™ PicoGreen® dsDNA Assay Kit (Invitrogen, Burlington, ON, Canada) with a VersaFluor fluorometer (BioRad, Mississauga, ON, Canada).
Polymerase chain reaction analyses
The primer sets used for PCR and the regions they amplified are described in Table 2. Primers for fragments F1 (62 bp), F2 (300 bp) and F3 (420 bp) were used to search for different sizes of transgenic DNA spanning the cp4 epsps construct in blood, digesta supernatant fractions, and batch-culture contents. The ovine-specific fragment GF (652 bp) was used as a positive control for amplification of DNA extracted from blood. The PCR conditions for F1 (62 bp) were: 95°C for 15 min; forty cycles of 95°C for 20 s and 60°C for 1 min; 72°C for 10 min. Conditions for F2 (300 bp), F3 (420 bp) and GF (652 bp) were: 94°C for 15 min; thirty-five cycles of 94°C for 30 s, 58°C for 30 s, 72°C for 1 min; 72°C for 10 min. Fragments F4 (1363 bp) and F5 (108 bp) were used to analyse transgenic DNA in whole RF and DF and faeces. PCR conditions for F4 were: 94°C for 15 min; 74°C for 5 min; thirty-five cycles of 94°C for 1 min, 74°C for 3 min; 72°C for 10 min. Conditions for F5 were: 50°C for 2 min; 95°C for 10 min; forty-five cycles of 95°C for 15 s and 60°C for 1 min. Universal primers were used as a positive control to detect bacterial DNA (fragment 16S; Table 2) encoding 16S from DNA extracted from RF and DF supernatant fractions, faeces, and batch cultures. Thermocycling conditions were: 94°C for 5 min; thirty cycles of 95°C for 15 s, 68·5°C for 30 s, and 72°C for 30 s, and an extension at 72°C for 10 min.
Table 2 Description of forward (F) and reverse (R) primers and a probe (P) used in the amplification of Roundup Ready® specific plant, bacterial, or ovine DNA

* Fragment F1 from Alexander et al. (Reference Alexander, Sharma, Deng, Whetsell, Jennings, Wang, Okine, Damgaard and McAllister2004); F2 and F3 from Sharma et al. (Reference Sharma, Alexander, John, Forster and McAllister2004); F4 from Alexander et al. (Reference Alexander, Sharma, Okine, Dixon, Forster, Stanford and McAllister2002); F5 from Monsanto Company (2005); 16S from Nadkarni et al. (Reference Nadkarni, Martin, Jacques and Hunter2002); growth factor from Lanneluc et al. (Reference Lanneluc, Mulsant, Harrison and Drinkwater1996).
† Transgene encoding the synthetic enzyme 5-enolpyruvylshikimate-3-phosphate synthase derived from Agrobacterium sp. strain CP4.
‡ Probe for real-time PCR.
Each conventional PCR mixture (50 μl) contained (final concentrations): 1 × HotStarTaq Master Mix (Qiagen Inc., Mississauga, ON, Canada), 0·15 μm each primer, 200 ng template DNA, with the exception of positive controls for blood (in which case 50 ng DNA was added to each PCR). The PCR were performed with a DNA Engine Dyad (M.J. Research Inc., Watertown, MA, USA). Following each PCR, 20 μl product was analysed on a 1·5 % (w/v) agarose gel.
Quantitative real-time PCR was performed using an iCyler iQ system (BioRad, Mississauga, ON, Canada) and primers amplifying fragment F5 (108 bp, Table 2) for detection of transgenic DNA in whole RF, whole DF and faeces. Each PCR (50 μl) contained (final concentrations): 1 × Universal Master Mix (Applied Biosystems, Foster City, CA, USA), 0·15 μm each primer and 0·05 μm probe. The probe (Table 2) was labelled at the 5′ end with the reporter dye 6-carboxyfluorescein (6-FAM) and at the 3′ end with the quencher dye, 6-carboxytetramethylrodamine (TAMRA). Standards were prepared using DNA isolated from RR rapeseed meal, and concentrations of 1·0 × 105, 2·0 × 104, 1·0 × 104, 2·0 × 103, 1·0 × 103, 200, 100 and 50 pg per reaction. As the amounts of transgenic rapeseed meal DNA standard were decreased, non-transgenic DNA was substituted in its place so that the final concentration of DNA (50 ng/μl) in each standard was kept constant. As well, DNA isolated from RF collected on the last day of period 1 from one sheep in each diet group, and pooled for extraction, was included with each standard. Each real-time PCR reaction contained 200 ng DNA.
Limits of detection of the 1363 bp cp4 epsps transgene
LOD assays were conducted to determine the sensitivity of the PCR for the 1363 bp cp4 epsps transgene (F4 from Table 2) in duodenal and ruminal matrices from sheep fed the concentrate-based or the forage-based diet. Four matrices were tested individually: 200 ng extracted DNA from RF or DF sampled from a single animal fed either the parental concentrate-based diet (animal 1) or the parental forage-based diet (animal 4) on day 0 were included in 50 μl PCR mixtures. Detection of the 1363 bp transgene was tested at concentrations of 1·0 × 104, 5·0 × 103, 1·0 × 103, 500, 200, 100, 50, 25, 12·5 and 0 pg purified RR rapeseed meal DNA per 50 μl reaction containing each matrix. Other PCR mixture components, thermocycling conditions and resolution of PCR products were identical to those described earlier for fragment F4 (1363 bp).
Statistical analyses
The amounts of transgenic rapeseed DNA measured by real-time PCR were analysed by PROC MIXED using the covariance structure UN (SAS/STAT version 8.1; SAS Institute, Cary, NC, USA; 1999). Differences were considered significant if the P values were less than or equal to an α value of 0·05.
Results and discussion
Transgene detection and quantification in whole duodenal fluid, ruminal fluid and faeces
Real-time PCR was used to measure the relative quantity of transgenic DNA in 2·0 × 105 pg DNA extracted from digesta samples. Transgenic DNA was quantifiable only in whole RF and DF, for up to 21 h after the last feeding of the forage-based or concentrate-based diets containing RR rapeseed meal (Fig. 2). Beyond 21 h, the amount of DNA in most RF and DF samples was less than the lowest standard (50 pg), which was considered the limit of quantification. Therefore, only DNA quantified between 1 and 21 h after feeding the last transgenic diets was statistically analysed.

Fig. 2 Quantification of transgenic DNA in whole ruminal fluid (○) and duodenal fluid (●) from sheep fed diets containing 15 % Roundup Ready® rapeseed meal. Values are the mean (n 6) DNA quantities, with standard errors represented by vertical bars. At 0 h, animals were fed diets containing transgenic rapeseed meal for the last time. At 24 h, animals were switched to diets containing 15 % parental rapeseed meal. The limit of quantification (····) was considered to be 50 pg. Statistical analysis was conducted only on data from time points 1 to 21 h.
Standard error was quite large as a result of variation among individual sheep. Diet type (forage-based v. concentrate-based) did not affect (P = 0·19) the relative quantity of transgenic DNA in RF or DF and therefore these data from animals fed the forage-based and concentrate-based diets were pooled to compare the effect of location (rumen v. duodenum) on transgenic DNA quantity (Fig. 2). There was a significant interaction between time of sampling and location of sampling within the digestive tract (P = 0·04; Fig. 2). The maximum mean amount of transgenic DNA occurred at 7 h after feeding in RF (1240 pg) and accounted for 0·62 % of total DNA added to the PCR (2·0 × 105 pg). Thereafter, the decline was steady, with the reduction attaining significance at 17 h (588 pg; P = 0·02). In contrast, the concentration of transgenic DNA in DF was more variable. Relative transgenic DNA content in DF was lower (P = 0·04) at 21 h (73 pg) than at 13 h (242 pg); however, differences in amounts of transgenic DNA among the other time points were not significant. The highest mean value for transgenic DNA in DF (360 pg) occurred at 7 h after feeding and represented 0·18 % of total DNA. The pattern of persistence of transgenic DNA in DF was similar to RF, but occurred later in the sampling period. This response probably reflects the transit time of rapeseed meal from the rumen to the duodenum. At 1, 4, 10, 13 and 21 h after feeding, the concentration of transgenic DNA was greater (P < 0·05) in RF than in DF. The DNA quantified in DF ranged from 7 % (at 1 h) to 35 % (at 13 h) of that quantified in RF. Rapeseed meal is a rapidly degradable protein supplement with digestion of 62·3 % DM after 12 h of incubation in the rumen (Piepenbrink & Schingoethe, Reference Piepenbrink and Schingoethe1998). While digestion of transgenic DNA within the rumen is apparently significant, it cannot be stated that 7 to 35 % of transgenic DNA from rapeseed meal passes from the rumen to the duodenum. This is because transgenic DNA was quantified within a fixed amount of total DNA (2·0 × 105 pg). Consequently, quantification is heavily dependent on the amount of microbial DNA extracted, which varies with time (Dehority & Tirabasso, Reference Dehority and Tirabasso2001) and location (Nicoletti et al. Reference Nicoletti, Davis, Hespell and Leedle1984) in the digestive tract.
In addition to transgenic DNA being quantifiable (>50 pg) up to 21 h after GM rapeseed was withdrawn from the diet, in some cases it was detectable beyond this time, as indicated by a positive cycle threshold during real-time PCR (Table 3). The 108 bp fragment F5 was detectable for 21 to 25 h in RF and 21 to 29 h in DF. The longer persistence in DF is probably due to passage time. At 25 h, the first meal of non-GM diets in period 3 had been consumed, probably diluting any remaining transgenic rapeseed meal, which in addition to its degradation and passage, rendered the transgene fragment undetectable shortly thereafter. We also investigated the detection of the 1363 bp cp4 epsps transgene (Table 3). This fragment was amplifiable from 1 to 13 h in RF, and from 4 to 10 h in DF. It has been theorised that transgenic plant DNA detected in a ruminal environment is limited to that protected within intact plant cells (Alexander et al. Reference Alexander, Sharma, Okine, Dixon, Forster, Stanford and McAllister2002). In that circumstance, one would expect that detection of one sequence of a plant genome would be as likely as any other, irrespective of size. However, there were clear differences in persistence of F4 (1363 bp) compared with F5 (108 bp). Initially, we thought that this could be a reflection of the LOD of F4 (1363 bp), due to its size, in RF and DF. However, results from LOD assays utilising DNA extracted from rapeseed meal showed this was not the case (Fig. 3). It appears, therefore, that despite being associated with solid plant material in RF and DF, the detection of transgenic DNA may not necessarily reflect protection within plant cell walls.
Table 3 Polymerase chain reaction detection of transgenic DNA in whole ruminal and duodenal fluids (RF; DF) of sheep fed diets containing 15 % Roundup Ready® rapeseed meal*
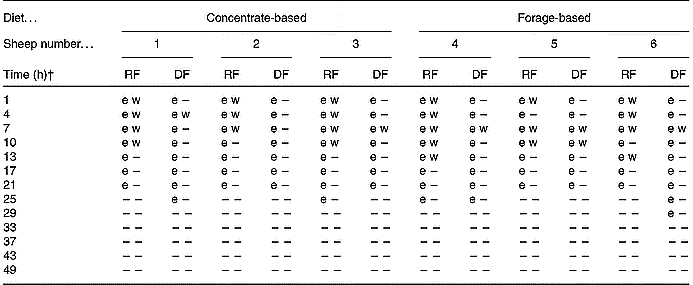
–, Not detectable.
* Detection of a 108 bp fragment spanning the 3′ insert-to-plant junction region (e) by real-time PCR or a 1363 bp fragment spanning the whole cp4 epsps gene (w) by conventional PCR. The fragments were considered positive when PCR resulted in a positive cycle threshold (e) for the 108 bp fragment or the proper band size when visualised by gel electrophoresis (w) for the 1363 bp fragment.
† Time point 0 h was considered to be 08.00 hours on day 11 of the trial when animals were fed diets containing Roundup Ready® rapeseed meal. At 08.00 hours on day 12 (i.e. time-point 24 h) and daily thereafter, the animals were fed diets containing parental rapeseed meal (see Fig. 1).

Fig. 3 PCR limits of detection of the 1363 bp 5-enolpyruvylshikimate-3-phosphate synthase transgene (cp4 epsps) in DNA extracted from ruminal (A, C) and duodenal (B, D) digesta of sheep fed a concentrate- (A, B) or a forage- (C, D) based diet. All PCR contained 200 ng matrix DNA from ruminal or duodenal digesta in addition to 10 000, 5000, 1000, 500, 200, 100, 50, 25, 12·5 or 0 pg extracted rapeseed meal DNA. L, DNA ladder.
The site of sample collection within the digestive tract influenced assay LOD. The LOD was lower in matrices from the rumen (50 pg) compared with the duodenum (100 pg). These results emphasise the importance of considering matrix effects in the detection of transgenic DNA in digesta contents. However, even when the amounts of transgenic DNA quantified were greater than the reported LOD (Fig. 2), detection of F4 (1363 bp) was not always possible. This may reflect differential digestion of plant DNA within the feed residue, which may be related to the nature of the bacteria involved in the digestion of feed particles. However, the present study would suggest that the differences in the microbial populations in sheep fed forage v. concentrate did not result in differential degradation of the transgene in rapeseed. Clearly, the detection of transgenic DNA in digesta samples is dependent on the selection of the fragment size to be amplified. Wiedemann et al. (Reference Wiedemann, Lutz, Kurtz, Schwarz and Albrecht2006) reported similar findings from ruminal in sacco incubation of transgenic maize. Although we reported the relative quantity of transgenic DNA based on amplification of a 108 bp DNA fragment (F5), the present results probably reflect the general trend of the fate of transgenic DNA during digestion of mixed diets containing rapeseed meal. It is clear that as digestion proceeds, the majority of transgenic DNA becomes increasingly reduced to a fragmented form, a phenomenon that favours the detection of smaller fragments.
From previous in vitro studies, transgenic DNA in diets containing GM rapeseed meal has been shown to be rapidly degraded within digesta (Flint & Thomson, Reference Flint and Thomson1990; Ruiz et al. Reference Ruiz, Andres and Smith2000; Alexander et al. Reference Alexander, Sharma, Deng, Whetsell, Jennings, Wang, Okine, Damgaard and McAllister2004; Sharma et al. Reference Sharma, Alexander, John, Forster and McAllister2004). In RF batch cultures, transgenic DNA could not be detected beyond 8 h of incubation. Further to these data, Wiedemann et al. (Reference Wiedemann, Lutz, Kurtz, Schwarz and Albrecht2006) described the ruminal digestion of transgenic maize DNA in sacco. Less than 10 % of the initial amount remained after 4 h of ruminal incubation (Wiedemann et al. Reference Wiedemann, Lutz, Kurtz, Schwarz and Albrecht2006). The present results demonstrate that transgenic DNA was quantifiable for up to 21 h in RF and DF (Fig. 2) and detectable for 21 to 29 h (Table 3). However, the present study does not imply that rapeseed DNA is stable in the rumen for that length of time, as feed consumption was in some cases protracted. Feed remained in the bunks of some sheep for up to 13 h after the morning delivery, despite being limited to 95 % of ad libitum intake. The disparity between these studies therefore reflects differences between the closed (in vitro and in sacco) systems and the in vivo circumstance studied here, in which ingestion of feed into the rumen occurred over a 6–13 h period.
Einspanier et al. (Reference Einspanier, Lutz, Rief, Berezina, Zverlov, Schwarz and Mayer2004) used real-time PCR to describe the persistence of transgenic DNA in the digestive tract of cattle fed GM maize silage, but were unable to reliably quantify transgenic DNA. However, they did measure a 97 % decrease in transgene copy number as a result of ensiling, which highlights the importance of initial gene copy in the feed for quantitative results. Fragments of DNA up to 23 kbp have previously been shown to be present in rapeseed meal and total mixed diets containing rapeseed meal (Alexander et al. Reference Alexander, Sharma, Okine, Dixon, Forster, Stanford and McAllister2002). Detection and quantification of transgenic DNA in vivo therefore depends on the quantity in feed (i.e. the proportion of GM plants in the diet), the extent of feed processing, the amount and time of feed intake, feed degradability, passage rates, dilution by microbial or endogenous plant DNA, and the fragment size of DNA being investigated.
None of the DNA extracted from faecal samples resulted in positive cycle thresholds, indicating the non-detection of fragment F5 (108 bp) in faeces. However, a 466 bp fragment of 16S DNA was amplifiable from each faecal sample (data not shown), confirming that the absence of detection was not due to PCR inhibition. This implies that transgenic DNA was substantially degraded during passage from the duodenum to the rectum, to levels at least as low as the LOD. Supporting these findings is a previous study in which transgenes from GM maize and soyabean were only detected in RF and DF of cattle, but not in faeces (Phipps et al. Reference Phipps, Deaville and Maddison2003). It also probably reflects the form in which GM rapeseed was fed to the sheep. Processing of rapeseed to meal and pelleting of the meal in a mixed diet significantly increased digestion of the DNA associated with the meal v. whole rapeseed, a result that reflects exposure of digestible cell components (i.e. cell wall, membrane and nucleic acids) to micro-organisms (Alexander et al. Reference Alexander, Sharma, Okine, Dixon, Forster, Stanford and McAllister2002). Passage of undigested whole rapeseed to the rectum would probably result in detection of transgenic DNA.
Transgene detection in digesta supernatant fraction and blood
Digesta samples were collected during period 2 (when forage- and concentrate-based diets containing GM rapeseed meal were fed) to investigate the fate of free transgenic DNA released from solid plant residue. Primers amplifying three fragments of DNA (F1, F2 and F3; Table 2) ranging in size from 62 to 420 bp and spanning the transgenic construct found in RR rapeseed were used. None of these fragments of transgenic DNA was detected in any of the RF or DF supernatant fraction samples collected approximately 4 h after feeding on days 3, 6, or 9 (data not shown). In contrast, detection of transgenic DNA in whole RF and DF was possible for up to 21 or 29 h after the last feeding of the GM diets (Table 3). It is likely though, that upon digestion, some free DNA is released into the aqueous digestive environment, as confirmed by detection of high-copy endogenous plant DNA in the liquid phases of RF and DF (Phipps et al. Reference Phipps, Deaville and Maddison2003). Evidence from an in vitro study indicates that such is also the case for transgenic DNA (Alexander et al. Reference Alexander, Sharma, Deng, Whetsell, Jennings, Wang, Okine, Damgaard and McAllister2004). In that report, however, detection of transgenic DNA was limited to small DNA fragments (62 bp) and the quantity of transgene in the DNA extracted from the feed residue was relatively high (approximately 2·5 × 104 to 3·5 × 104 copies per 245 ng DNA). In the present study, the average amount of transgenic DNA extracted from whole RF or DF did not exceed 1240 or 367 pg, respectively, in 2·0 × 105 pg of extracted DNA. Any free transgenic DNA released into the rumen was probably rapidly degraded to below detectable levels. The fact that transgenic DNA was not detected in the aqueous phase of RF from either diet suggests that abundant microbial nucleases were present to rapidly degrade DNA in both concentrate- and forage-based diets. Negative results for transgenic DNA in the DF supernatant fraction were probably not a result of enzymic degradation of DNA, however, as the pH of DF is too low for the optimal activity of such enzymes (Armstrong & Hutton, Reference Armstrong, Hutton, McDonald and Warner1974). All of the supernatant fraction samples did contain quantifiable DNA, most of which was probably microbial. A 466 bp fragment of 16S DNA was amplifiable from each sample, indicating the presence of bacterial DNA. This therefore suggests that the inability to detect transgenic DNA in RF and DF supernatant fractions did not result from PCR inhibitors.
The same fragments of DNA (F1, 62 bp; F2, 300 bp; F3, 420 bp) sought for amplification in cell-free digesta supernatant fractions were analysed in DNA extracted from blood. In addition, blood was analysed for fragment F5 (108 bp) by real-time PCR. The ovine growth-factor gene (positive control) was amplifiable in all of the blood samples tested, confirming that extracted DNA was of sufficient quality for PCR (Fig. 4(A)). However, none of fragments F1 (62 bp), F2 (300 bp) or F3 (420 bp) was visible by gel electrophoresis (Fig. 4(B)) after PCR of any of the DNA samples, nor did real-time PCR yield a positive cycle threshold for F5 (108 bp; data not shown).

Fig. 4 Gene detection in DNA extracted from whole sheep blood while being fed diets containing 15 % GM rapeseed meal. Animals A1, A2 and A3 were fed a concentrate-based diet and A4, A5 and A6 were fed a forage-based diet. (A) Detection of an ovine-specific growth factor gene in blood on days (d) 1, 5 and 9. L, DNA ladder; Neg, negative control containing all components of the PCR except template DNA. (B) Representative analyses for PCR of transgenic fragments in blood from A1 and A4 on d1 and 5. All blood samples tested negative for 62 bp, 300 bp and 420 bp fragments of transgenic DNA. Pos, positive control containing DNA extracted from Roundup Ready® rapeseed meal.
There is evidence that absorbed DNA may be associated with leucocytes (Schubbert et al. Reference Schubbert, Renz, Schmitz and Doerfler1997; Einspanier et al. Reference Einspanier, Klotz, Kraft, Aulrich, Poser, Schwagele, Jahreis and Flachowsky2001). Therefore, DNA was extracted from whole blood so as to include DNA associated with both blood cells and plasma. To cross the intestinal barrier, transgenic DNA would probably have to exist in a free form in the fluid phase of digesta. The non-detectability of free transgenic DNA in the aqueous phase from DF suggests that release of DNA fragments must have occurred distal to the duodenum to enable their absorption across the intestinal epithelium. The present study confirms that transgenic DNA did reach the small intestine (Table 3), and subsequently was degraded to below detectable levels, as fragments of the transgene were not detected in the faeces. Whether or not transgenic DNA did exist in free form, separate from solid feed residue, between the duodenum and rectum, is unknown. As mentioned earlier, the average amount of transgenic DNA extracted from whole DF did not exceed 367 pg in 2·0 × 105 pg total DNA (approximately 0·18 %). The substantial dilution of transgenic DNA in non-transgenic DNA would limit its probability of absorption. Plant DNA does apparently cross the intestinal epithelium, as evidenced by fragments of endogenous high-copy plant DNA detected in the blood (Einspanier et al. Reference Einspanier, Klotz, Kraft, Aulrich, Poser, Schwagele, Jahreis and Flachowsky2001), muscle (Nemeth et al. Reference Nemeth, Wurz, Artim, Charlton, Dana, Glenn, Hunst, Jennings, Shilito and Song2004) and milk (Nemeth et al. Reference Nemeth, Wurz, Artim, Charlton, Dana, Glenn, Hunst, Jennings, Shilito and Song2004; Phipps et al. Reference Phipps, Jones, Tingey and Abeyasekera2005) of cattle. The extent of this uptake is probably dependent, however, on the number of copies of a gene found in feed, which itself is directly related to the gene copy number within the plant's genome and concentration of GM feed in the diet. Low-copy genes have not been reported in ruminant products, to the best of our knowledge.
Persistence of transgenic DNA in batch cultures
It is difficult to determine whether or not bacteria in RF and/or DF are being transformed by transgenic DNA because of the confounding effect of contamination by the transgenic plant material itself. Thus, the decision was made to subculture the microbes in whole RF and DF collected during period 2 (i.e. during intake of RR rapeseed meal) to test for stable incorporation of transgenic DNA. All of the RF and DF used as inoculum in the initial batch-culture incubation tested positive for plant fragments F1 (62 bp), F2 (300 bp) and F3 (420 bp) and for bacterial fragment 16S (466 bp; data not shown). After 54 h of subculturing (3 × 18 h), however, none of these transgenic plant DNA fragments was amplifiable in the batch-culture incubation liquids, whereas fragment 16S (466 bp; bacterial control) was still detectable (data not shown). This suggests that the transgenic DNA fragments detected in RF and DF were associated with suspended plant material, and not with DNA within the microbial genome. Alterations in mixed microbial populations in the gut are known to occur in response to changes in the diets fed to ruminants, for example, feeding grain v. hay (Tajima et al. Reference Tajima, Aminoy, Nagamine, Matsui, Nakamura and Benno2001). Regardless of the diet type in the present study, stable incorporation of transgenic DNA into microbial genomes did not occur at detectable levels in batch cultures. The same is true for fragments F4 (1363 bp) and F5 (108 bp) in the microflora of RF and DF, as shown by the finite intervals during which the fragments were detectable (Table 3). Low-copy transfer of transgenic plant DNA to gut microflora of human subjects has been described (Netherwood et al. Reference Netherwood, Martìn-Orúe, O'Donnell, Gockling, Graham, Mather and Gilbert2004). In ruminants, naked plasmid DNA has been shown to lose the capacity to transform bacteria within 1 min of incubation in RF (Duggan et al. Reference Duggan, Chambers, Heritage and Forbes2000). Similarly, the present study demonstrated non-detectability of free transgenic DNA from rapeseed meal in cell-free RF and DF supernatant fractions, implying that the instability of plant DNA in digesta is an important limitation to the likelihood of introduction of the transgene into microbial genomes. Additionally, dilution of transgenic DNA probably reduces the likelihood of this process. At its peak concentrations in whole RF and DF, transgenic DNA accounted for only 0·54 and 0·18 % of the total DNA, respectively. It remains to be determined if the microflora that are closely attached to feed particles are exposed to free plant DNA for an extended period of time.
Conclusion
In conclusion, the present study revealed that the disappearance from the rumen of transgenic DNA in diets containing 15 % RR rapeseed meal results from ruminal digestion and passage to the small intestine. This was indicated by detection, in the duodenum, of genes as large as 1363 bp. However, transgenic DNA located in the duodenum is degraded to such an extent that it escapes detection by the time it passes to the rectum of sheep. The extremely low presence (undetectable) of free plant DNA in the supernatant fractions from RF and DF is a limiting factor in the transformation of microbes and absorption across the intestine. Evidence of either of these events was not found. The dilution of transgenic DNA amongst other plant and microbial DNA is an additional impediment to its uptake by bacteria or host animal. Quantitative real-time PCR can be used to investigate the fate of plant DNA in ruminants, but this may be limited by the starting quantity of DNA in the feed and its digestion. Quantification of genes specific to individual plants may allow comparison of their digestion in the rumen, given that the majority of plant DNA appears to be associated with solid feed residues. However, the nature of this association requires further study as it is not clear whether plant DNA is encased within plant cells or associated with the feed matrix. Detection of transgenic DNA in livestock is dependent on a number of factors, including matrix effects, LOD, quantity of transgene in the feed, digestibility of the feed, and differences in the digestive physiology among livestock.
Acknowledgements
The authors would like to thank Monsanto Company for providing Roundup Ready® rapeseed meal and event-specific primers and probes for detection purposes. Thanks are also extended to staff at the sheep barn, Ken Wright and Patrick Kubik for their help.









