INTRODUCTION
Yersinia pestis, a zoonotic bacterium that is the causative agent of plague, is increasingly recognized as a re-emerging threat to human and animal health [Reference Prentice and Rahalison1]. The disease is widely endemic [Reference Stenseth2]; worldwide, about 1000–5000 cases and 100–200 deaths per year were reported between 1987 and 2001 [3]. In total, 24 countries reported cases in this time period, including from Africa, Asia and the Americas. Because detection and reporting are probably limited in most endemic areas, this is likely to be an underestimation.
During epizootic outbreaks, the main mode of transmission is from animals to humans through the bite of infective fleas exposed to the main zoonotic reservoir(s), which are small mammals and rodents. More rarely, transmission can occur from larger mammals (e.g. camels) or through direct contact with an infectious source or inhalation of infectious respiratory droplets. Plague typically manifests in three forms – bubonic, septicaemic, and pneumonic, depending on the route of infection [Reference Worsham, Lenhart, Lounsbury, Martin and Dembek4]. Clinical features vary depending on the form, but generally include fever, chills, malaise, hypotension, lymphadenopathy; fatality rates of 100% in untreated cases have been reported [Reference Worsham, Lenhart, Lounsbury, Martin and Dembek4]. Plague is a notifiable disease and the bacterium is considered as a potential bioterrorism agent. Although rare, cases of plague have been reported after the consumption of camel meat [Reference Arbaji5–Reference Christie, Chen and Elberg7]; however, it can often be difficult to determine whether the infection occurred from the consumption of the infected meat or by direct contact with the infectious animal, and/or its fleas, during slaughter [Reference Christie, Chen and Elberg7]. While plague remains endemic in parts of Asia, Africa, and the USA, it has never been reported in Afghanistan [Reference Stenseth8].
Detecting and responding to outbreaks of acute diseases remains a challenge in low-resource settings, where systems, expertise and resources are lacking. In Afghanistan, there is a range of endemic infectious diseases [Reference Wallace9]. A nascent disease surveillance system – the Disease Early Warning System (DEWS) – is currently the formal system for detection of outbreaks. Reports of unusual outbreaks or spikes in the number of cases presenting at clinics are reported from clinics, hospitals and communities to the DEWS. The system is based predominantly on syndromic descriptions for diagnosis of disease because laboratory capacity in Afghanistan is insufficient for reliable identification of rare infectious diseases.
Reports of an outbreak of acute gastroenteritis which included deaths were received by DEWS from the provincial hospital in the southwestern Afghan province of Nimroz in December 2007. The symptoms reported were fever and severe gastroenteritis with rapid onset, in some cases including lymphadenopathy and more rarely, pharyngeal lesions. As reported by hospitalized victims, and confirmed by the community, just prior to the recognition of cases a camel that was clinically ill was slaughtered and distributed among many residents of one village and to some residents of two neighbouring villages. The animal was reported to be visibly ill, in retrospect with an infectious disease, exposing up to 2000 people potentially. Because of this and the severity of symptoms, an outbreak investigation was undertaken. The spectrum of clinical features, and the potential involvement of camel meat, led to an initial hypothesis that Bacillus anthracis was the responsible pathogen. Human cases of gastrointestinal anthrax associated with infected livestock have been widely reported in the past [Reference Sirisanthana and Brown10].
We describe the epidemiological findings of the outbreak investigation and the molecular detection of Y. pestis from human clinical specimens and camel meat.
METHODS
Study site, epidemiological investigation, and case definition
We conducted an epidemiological investigation consisting, first, of active case detection in three villages, and second, of a cross-sectional survey in two of the affected villages. The three villages were located in Kang district, Nimroz province of southwestern Afghanistan. This province is near the Iranian border and is in an arid desert landscape.
Active case detection involved approaching community leaders and gaining their assistance in surveying the villages for any person who fitted the characteristics of the outbreak – any severe gastroenteritis or associated symptoms; we also enquired if patients had any contact with the camel, and if so, what type of contact (e.g. slaughter, cooking, etc.). Hospitals and clinics in the area and provincial capital were also surveyed for patients from the villages and with a history of symptoms consistent with other cases. Cases were followed up in their homes (or in the clinic or hospital). The number of cases with history of fever, diarrhoea, vomiting, pharyngeal lesions, lymphadenitis and other associated symptoms were noted and reports of any deaths were collected including interviews with close family members in order to ascertain symptoms. Cases were treated with ciprofloxacin, referred to hospital if necessary, and prophylaxis, also with ciprofloxacin, was provided to household members of cases.
For the second phase (the cross-sectional survey), data were systematically collected from all households in the most affected village and a random selection of about 50% of households in the second affected village. Houses were approached and verbal consent was received from all participants, after which trained field surveyors administered a structured questionnaire. This provided information on symptoms, treatment and potential exposure factors. A wide range of symptoms were recorded, but the clinical picture from cases in hospitals included fever, vomiting, diarrhoea and in some cases pharyngeal lesions and lymphadenitis as the primary descriptive symptoms. Exposure factors included in the questionnaire were: food consumption history (from the date of the camel's slaughter) including normal staple foods (rice, bread, vegetables) and meats; water source; contact with the slaughtered camel, including participation in the slaughter, processing of the carcass, handling and cooking of meat and the presence of other animals at home. Data were noted on the forms by the surveyors and then double-entered using Microsoft Access. Data were analysed using Stata version 8 (StataCorp, USA).
At the time of the epidemiological investigation there was no confirmed causative agent so case definitions were based on the primary symptoms of the outbreak. For the epidemiological investigation, we used two definitions of a case. A suspected case was defined as having any of fever, vomiting, diarrhoea, or abdominal pain since the day of the camel's slaughter; and probable cases were defined as having all of fever, vomiting and diarrhoea over the same time period. Since bacterial culture and identification capacity was low, definitive diagnoses remained unavailable at the time of the survey.
To test the primary hypothesis that the source of the outbreak was the camel, we examined a range of pre-defined exposure factors against the two clinical outcomes (the case definitions). As a primary analysis, individual potential risk factors were cross-tabulated using χ2 tests for association with each of the outcomes (both case definitions). Multivariate logistic regression analysis was then conducted using the same predefined exposure variables to adjust for potential confounding between them and the outcome.
Laboratory analysis
Seven whole-blood and serum samples were collected from three hospitalized patients. Whole blood was cultured locally. All seven samples were subjected to ELISA testing for evidence of Congo-Crimean haemorrhagic fever (CCHF) and B. anthracis at the United States Army Medical Research Institute of Infectious Diseases (USAMRIID).
Parts of the camel were located and exhumed. Specimens (skeletal muscle) were extracted and transported to Kabul. Bacterial cultures of the camel meat were conducted at the Central Public Health Laboratory in Kabul, Afghanistan.
Subsequently, sera and whole blood from three of the non-fatal human cases (patients 1–3), along with samples of the remaining camel meat were analysed using broad-range PCR/electrospray ionization–mass spectrometry (PCR/ESI–MS), followed by real-time PCR. DNA was extracted from all patient samples and camel meat using the QIAamp DNA extraction kit (Qiagen, USA) according to the manufacturer's instructions. Extracted DNA from all samples was analysed using PCR/ESI–MS on the Ibis T5000 platform. This system is a multiplexed detection format which uses broad-range PCR followed by specific pathogen identification by mass spectrometry to determine the amplicon's unique base composition signature [Reference Ecker11]. The specific assay used for this analysis was the Biodefense Bacterial and Viral Surveillance kit (Abbott Molecular Inc., USA), which targets 13 bacterial and four viral biothreat agents and distinguishes them from their near neighbours in a single assay format.
Confirmatory testing was performed using three LightCycler (Roche Diagnostics, USA) real-time PCR assays previously developed for detecting Y. pestis [Reference Christensen12]. These assays specifically target the genes for the pesticin immunity protein (pim), plasminogen activator (pla), and the capsular antigen F1 (caf1), which are known virulence factors specific for Y. pestis. IgG and IgM enzyme-linked immunosorbent assays (ELISA) were performed on patient sera as previously described [Reference Duermeyer, Wielaard and van der Veen13, Reference Iacono-Connors14]. Both ELISAs were designed to detect antibodies specific for the Y. pestis F1 antigen. Titres reported are equal to the reciprocal of the last dilution that is above or equal to the optical density cut-off value, and samples were considered positive if their titre was ⩾100.
RESULTS
Epidemiology
The outbreak was reported over 15 days in the villages following the slaughter of the camel on 16 December 2007. Camel meat had been distributed to 150–200 families. Fifty cases were found in the active case-detection phase of the investigation, with eight reported deaths all of which came from the village where the camel was slaughtered (Burrida). These deaths reportedly included three members of the same family who had slaughtered the camel, including the butcher.
A total of 607 persons were enrolled in the cross-sectional survey and included 382 suspected cases and 83 probable cases (Table 1). All residents of Burrida (comprising 30% of the sample) and 50% of the second village (Shandro) were surveyed.
Table 1. Enrolment characteristics of the cross-sectional survey sample
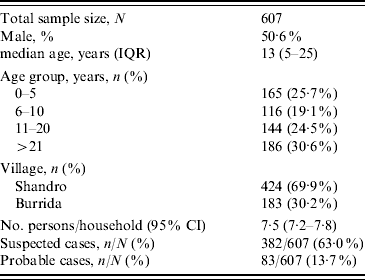
IQR, Interquartile range; CI, confidence interval.
Of the probable cases, 17 had died with reported symptoms consistent with the disease, giving an estimated case-fatality rate of 20·5%. Exposure variables were examined by univariate and multivariate analysis against each of the case definitions. Among the food consumption risk factors, eating camel meat was the only factor positively associated with being a case. Of those who had consumed camel meat, 253/347 (72·9%) were suspected cases vs. 129/260 (49·6%) in those who had no history of consumption (χ2=34·6, P<0·001). Consumption of cow meat (beef) was negatively associated with disease in suspected cases [84/225 (37·3%) in non-cases vs. 83/382 (21·7%) in suspected cases; χ2=17·3, P<0·001]. This association is probably because those who consumed cow meat (beef) were less likely to have consumed camel. Of those who consumed camel meat, 64/347 (18·4%) were probable cases vs. 19/260 (7·3%) who had no history of consumption (χ2=15·6, P<0·001).
Other associated exposure factors for suspected cases were cooking camel meat: 79/92 (85·9%) were cases vs. 303/515 (58·8%) who did not cook camel meat (χ2=24·5, P<0·001), although there was no association with these two factors and being a probable case. Any exposure to camel meat, through eating, handling, cooking or slaughter was associated with being a suspected case and a probable case (Table 2). No association was found with other foodstuffs or water source. Age group was negatively associated with being a probable case (adjusted odds ratio 0·6, 95% confidence interval 0·5–0·8).
Table 2. Multivariate logistic regression analysis of potential pre-specified risk factors against the two primary outcomes in the study
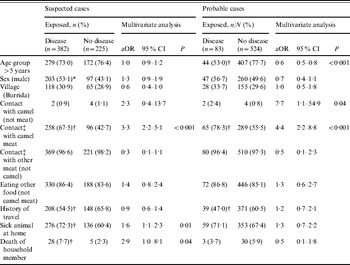
aOR, Adjusted odds ratio, using pre-specified potential confounding factors in a logistic regression model; CI, confidence interval.
* χ2: P<0·05.
† χ2: P<0·01.
‡ Eating, handling, cooking, and/or slaughter.
Laboratory confirmation
The initial bacterial culture of camel meat, conducted in Kabul, was inconclusive and although it yielded bacilli, they could not be identified to species level. In retrospect these probably represented soil contaminants since the meat had been buried prior to being exhumed for submission to the laboratory. Blood culture of patients produced no growth; however, patients had been treated with ciprofloxacin prior to sampling which may have cleared any bacteraemia.
After the outbreak had concluded, in May 2008 samples of blood (n=2) (acute samples from patients 1 and 2) and sera (n=3) (convalescent samples from patients 1–3) were tested at USAMRIID. The initial ELISA tests for Bacillus anthracis and CCHF virus, as well as bacterial and viral culture attempts, were uniformly negative.
Three patients and one sample of camel meat subjected to further analysis with the PCR/ESI–MS assay detected the presence of DNA signatures consistent with Y. pestis in both an acute whole-blood specimen from patient 1 and in the camel tissue (Fig. 1a, b). All three Y. pestis-specific primer pairs that are incorporated into the PCR/ESI-MS assay, targeting the inv, pla, and caf genes, were positive for both samples and produced identical base composition signatures in the patient specimen and the camel tissue (Fig. 1c). Y. pestis DNA was not detected in blood or sera from patients 2 and 3.

Fig. 1. PCR/ESI–MS analysis of patient and camel samples. Spectra of the amplicons resulting from PCR targeting the Y. pestis caf gene from an (a) acute whole-blood specimen collected from patient 1 and (b) camel meat. (c) Base compositions (A, T, G, C) resulting from PCR of the Y. pestis-specific genes, inv, pla, and caf showing identical base compositions within the gene targets between the two samples.
As further confirmation of the presence of Y. pestis is these samples, we used three LightCycler (Roche Diagnostics) real-time PCR assays previously developed for detecting Y. pestis. These assays target the virulence factor genes, pim, pla, and caf, of Y. pestis. As with the PCR/ESI–MS analysis, real-time PCR testing targeting all three genes showed that the acute whole-blood specimen from patient 1 and the camel tissue were strongly positive for the presence of Y. pestis (Table 3). Furthermore, acute blood and convalescent sera (taken ~1 month after the outbreak) from patients 1 and 2 and convalescent serum from patient 3 were tested for the presence of IgM and IgG antibodies specific to the F1 antigen of Y. pestis using ELISAs. While patient 1 did not have any antibodies to Y. pestis, convalescent serum of patient 2 had both IgM and IgG anti-F1 antibodies (Table 3). The acute serum sample from this patient was negative, providing evidence for seroconversion and, thus, recent infection with Y. pestis. Convalescent sera from patient 3 also had positive IgG titres to F1; however, because it was IgM negative and no acute sample was available, we cannot rule out the possibility that this result represents antibody from a previous exposure.
Table 3. Summary of diagnostic testing on patient specimens and camel tissue from the 2007 Afghan outbreak
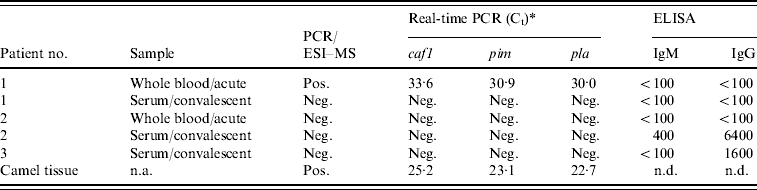
caf1, Capsular antigen F1; pim, pesticin immunity protein; pla, plasminogen activator; n.a., not applicable; n.d., not determined.
* Crossing point.
DISCUSSION
In humans, infection with Y. pestis typically occurs from the bite of an infected flea and manifests in one or more of three clinical forms: bubonic, septicaemic, or pneumonic. We describe the epidemiology of an outbreak of severe gastrointestinal disease in Afghanistan after the consumption of camel meat in late 2007. We provide laboratory evidence that this was a rare outbreak of gastroenteritis caused by the bacterium Y. pestis, which probably resulted from the consumption of infected camel meat. Those who had contact with the camel were also at elevated risk, but the primary public health concern was for the widespread distribution of the camel meat where several hundred individuals were found (in our sample) to be exposed. We were not able to identify the infectious agent until several months after the outbreak but the epidemiological data strongly implicated consumption and exposure to camel meat as the route of transmission. Because the outbreak was limited in time, the response focused on treatment of those affected using active case detection. Plague was not identified in time to put in place conventional control mechanisms (flea and rodent control), although this would not have been appropriate because of the unusual route of transmission.
While human plague is most often acquired from the bites of infected fleas, rare sporadic cases due to exposure to infected carcasses of camels and other animals have been documented [Reference Wernery and Kaaden15]. In particular, in more recent times, the consumption of camel meat was implicated in human plague cases from, Jordan [Reference Arbaji5], Saudi Arabia [Reference Bin Saeed, Al-Hamdan and Fontaine6] and Libya [Reference Christie, Chen and Elberg7]. Although no severe pharyngitis, tonsillitis or pharyngeal lesions were observed in the Afghan patients, gastrointestinal symptoms such as vomiting, diarrhoea, and abdominal pain suggested ingestion as the route of exposure. Outbreaks of this nature are rare, but are often severe in nature. The infected camel in this case reportedly had an open, ulcerated lesion on its hock, so it seems likely that it contracted plague from the bite of an infected flea somewhere along the route of its journey, possibly days to weeks before the outbreak. It was not possible to track the camel's journey with any accuracy, so the possible location of infection could not be ascertained. Once infected, the camel had become clinically ill but was slaughtered and the meat distributed widely. Cooking practices may explain differential exposure and thorough cooking almost certainly would have limited the outbreak. A speciality of rare-cooked camel is evident in this region, and while some may have prepared the food in this way, most of it would have been cooked thoroughly. Those who were closely associated with the slaughter and handling of the camel meat, including cooking, had elevated risk of disease and death. The attack rate in patients who had cooked the meat was ~80%.
Detection of the outbreak via the Afghanistan DEWS allowed a rapid response to be mounted. Part of the response was to conduct the investigation and to ensure that patients were adequately treated. The disease was correctly presumed to be infectious by the treating physicians, and specifically bacterial. Thus, cases were appropriately treated with ciprofloxacin and appeared to respond. In light of the rarity of plague as the cause of such an outbreak, diagnosis of the infectious agent would have been challenging under any circumstances, and was more of a challenge in the context of Afghanistan's reconstruction. Thus, the working hypothesis that the outbreak was the result of anthrax was not disproved until several months after the initial outbreak. This identifies the challenge of detecting infectious agents in low-resource settings if the disease is uncommon and case presentation is unusual.
All bacterial culture attempts from patients' blood and camel tissue were negative or only yielded contaminants, and few samples were available for testing; the key to identifying the causative agent in this outbreak was the use of the Ibis T5000 PCR/ESI–MS assay. This diagnostic tool has previously been shown to detect a broad range of bacterial pathogens [Reference Baldwin16, Reference Postinikova17] and several viruses [Reference Eshoo18–Reference Sampath20]. The inclusion of certain key gene targets in the Ibis T5000 biodefence assay allows it to detect and identify a wide range of pathogens in potentially complex clinical or environmental samples with little or no background effect. Specifically, three of the gene targets included in this multiplexed assay are the pim, pla, and caf genes of Y. pestis. All three of these targets were positive in the acute whole-blood specimen from patient 1 and the camel tissue (Fig. 1c). Follow-up testing using real-time PCR confirmed these results, and immunological testing indicated seroconversion to Y. pestis F1 antigen in patient 2.
The presence of Y. pestis has not been reported from Afghanistan previously, despite reports from neighbouring countries [Reference Stenseth8]. A number of zoonotic diseases causing acute outbreaks of disease threaten the inhabitants of Afghanistan and the surrounding region, including CCHF and now plague. Thus, enhanced disease surveillance efforts and improved laboratory diagnostic capabilities are required to mitigate the effects of outbreaks of acute diseases.
CONCLUSIONS
In summary, we described the first reported occurrence of plague in Afghanistan. In this outbreak of disease, illness was strongly associated with consumption of camel meat. Using epidemiological analysis and a novel PCR/ESI–MS assay, real-time PCR, and immunological analysis of diagnostic specimens from both patients and camel tissue, we provided strong evidence that the 2007 Afghanistan outbreak was due to plague and that consumption of Y. pestis-infected camel meat was the source of the infection. Enhanced detection and surveillance mechanisms, including improved laboratory diagnosis capacity, is required for identification of rare outbreaks of acute disease in developing countries.
ACKNOWLEDGEMENTS
We thank Tamara Clements for conducting the ELISAs. Laboratory research was funded, in part, by the Defense Threat Reduction Agency, Fort Belvoir, VA. Fieldwork was funded by WHO Kabul and United States Naval Medical Research Unit no. 3, Cairo, Egypt. The opinions, interpretations, conclusions, and recommendation are those of the authors and are not necessarily endorsed by the U.S. Army or any other associated agency.
DECLARATION OF INTEREST
None.






