In recent years, our knowledge on the composition of the human intestinal microbiota has greatly improved with the introduction of molecular techniques based on 16S rRNA gene sequences, confirming that the microbiota is subject specific and comprises a complex and very diverse bacterial ecosystem of which most species belong to previously unknown (i.e. uncultivated) lineages(Reference Rajilic-Stojanovic, Smidt and de Vos1–Reference Zoetendal, Vaughan and de Vos3). In healthy individuals, the dominant intestinal microbiota is found to be relatively stable over time(Reference Rajilic-Stojanovic, Smidt and de Vos1, Reference Maukonen, Matto and Satokari4–Reference Zoetendal, Rajilic-Stojanovic and de Vos7). However, in various populations it has been demonstrated that antibiotic intake can cause a marked disturbance of the intestinal microbiota as they not only affect pathogens but also the indigenous microbiota, which may result in antibiotic-associated diarrhoea (AAD). It is widely assumed that this disturbance is short-term but recently also medium and long-term disturbances in (specific) bacterial populations have been described(Reference Dethlefsen, Huse and Sogin8–Reference Koning, Jonkers and Stobberingh10). However, there are chronically ill patients, such as patients suffering from chronic obstructive pulmonary disease (COPD), who receive frequent antibiotic therapy. COPD is characterised by a progressive airflow obstruction, associated with a chronic inflammatory response in the airways consisting mainly of neutrophils, CD8+T-cells and macrophages(Reference Barnes11, Reference Turato, Zuin and Saetta12). Acute exacerbations play an important role in the clinical course of the disease and are an important cause of morbidity and mortality(Reference Burge and Wedzicha13, Reference Donaldson, Seemungal and Patel14). Although the underlying pathogenic mechanisms are poorly understood, antibiotic therapy is frequently prescribed. The effects of such frequent antibiotic use on the intestinal microbiota are, to our knowledge, not known.
The intestinal microbiota plays an important role in human physiology, being involved in gut maturation, colonisation resistance, several metabolic processes, regulation of intestinal epithelial proliferation and modulation of the mucosal and systemic immune response(Reference Cebra15–Reference Noverr and Huffnagle18). Based on this, probiotics are often given during and after antibiotic intake and beneficial effects have been observed in the prevention and treatment of AAD(Reference Cremonini, Di Caro and Nista19–Reference McFarland22). Therefore, in the present study a multispecies probiotic was given containing nine different probiotic strains selected on the basis of their in vitro ability to inhibit growth of Clostridium spp., in vitro survival of gastrointestinal passage(Reference Timmerman, Niers and Ridwan23) (i.e. low pH (2·5) as well as bile and digestive enzymes (pancreatin and pepsin)) and the absence of acquired antibiotic resistance. Moreover, a combination of strains were selected in which strain-specific properties were chosen to be additive or synergistic and mutual inhibition was absent. In addition, we previously showed that in healthy volunteers the intake of this multispecies probiotic during and after amoxycillin intake has no adverse effects and affects the faecal microbiota resulting in a faster restoration toward the pre-antibiotic state(Reference Koning, Jonkers and Stobberingh10).
The aim of the present study was to assess the disturbance of the dominant faecal microbiota and the possible restoration by a multispecies probiotic in patients with COPD treated with antibiotics for an acute exacerbation. Moreover, the effect on specific bacterial subgroups, using both culture-dependent and molecular based techniques, bowel habits, the incidence of antibiotic resistance, endotoxins and pH was also studied.
Methods
Subjects
Forty-five patients, 18–80 years of age, with moderate to severe COPD admitted to the Centre for Integrated Rehabilitation of Organ Failure (Horn, The Netherlands) for a comprehensive interdisciplinary pulmonary rehabilitation program were included in the study. Patients had to have an acute exacerbation of COPD, which according to the Anthonisen criteria(Reference Anthonisen, Manfreda and Warren24) should be treated by a 7 d antibiotic regimen, as judged by the physician in charge. Exclusion criteria were: immune-compromised subjects or treatment with immune-suppressive medication other than for COPD, pregnancy, lactation, hypersensitivity to the commonly prescribed antibiotics, pre-existing bowel pathology (including irritable bowel syndrome, inflammatory bowel disease, diverticulitis and cancer) and diarrhoea or constipation (in the last 3 d before inclusion). When using doxycycline maintenance therapy, corticosteroids or gastric acid inhibitors, patients had to be on stable medication for more than 2 weeks. Furthermore, patients were not allowed to use (1) laxatives and anti-diarrhoea medication in the 2 weeks before the start of the study and during the study; (2) regular pro- and prebiotic intake in the 2 weeks before the start of the study and during the study. Finally, ex-smokers had to have quitted smoking for at least 2 weeks before the start of the study.
The present study was conducted according to the guidelines laid down in the Declaration of Helsinki and all procedures involving patients were approved by the Medical Ethics Committee of the University Hospital Maastricht, The Netherlands. Written informed consent was obtained from all the patients.
Study design
The study was executed according to a parallel, randomised, placebo-controlled, double-blind design. The total duration of the intervention and follow-up period was 63 d. All the patients were prescribed a 7 d antibiotic treatment and were randomised to receive either 5 g of a multispecies probiotic, Ecologic® AAD, or placebo twice daily for 14 d starting simultaneously with antibiotic treatment. This resulted in three time periods defined as: days 1–7, ‘the antibiotic/probiotic period’ in which all patients received a 7 d antibiotic treatment in combination with either probiotic or placebo; days 8–14, ‘the probiotic only period’ in which patients received either probiotic or placebo; days 15–63, ‘the post-treatment follow-up period’. The antibiotic treatment was given according to the physician's instructions (care as usual). The time between antibiotic and probiotic intake had to be at least 2 h. Fresh faecal samples were collected on days 1 (the first sample patients were able to produce after inclusion), 7, 14 and 63. On the same day, a questionnaire was filled out including the clinical COPD questionnaire(Reference van der Molen, Willemse and Schokker25), questions on bowel movements (stool frequency and consistency (ranging from ‘1’, hard lumps, to ‘7’, completely watery) according to the Bristol stool form scale(Reference O'Donnell, Virjee and Heaton26)), use of pre- and probiotics, other medication taken, change of eating habits and compliance. In addition, a short questionnaire on bowel habits and side-effects (nausea, abdominal cramps, bloating, flatulence or other) had to be completed daily during probiotic/placebo intake. Moreover, this questionnaire was used to determine diarrhoea-like defecation defined as a defecation frequency ≥ 3/d and/or a faecal consistency (on the Bristol stool form scale) ≥ 5/d for at least 2 d.
Probiotic
The multispecies probiotic (Ecologic® AAD) and the placebo were kindly provided by Winclove Bio Industries, Amsterdam, The Netherlands. Ecologic® AAD consists of nine different bacterial species at 108 colony forming units/g each, 5 % mineral mix and 15 % Raftilose® Synergy1. All strains are deposited in the Dutch Dairy Institute (NIZO, Ede, The Netherlands) culture collection and are characterised with (GTG)5-PCR fingerprinting as well as further fingerprinting using BOX-A1R and OPY11 primers, followed by SSU (16S)-rRNA sequence analysis (Bifidobacterium bifidum NIZO 3804, Bifidobacterium lactis NIZO 3680 (formerly known as Bifidobacterium longum), Enterococcus faecium NIZO 3886, Lactobacillus acidophilus NIZO 3678 and NIZO 3887, Lactobacillus paracasei NIZO 3672, Lactobacillus plantarum NIZO 3684, Lactobacillus rhamnosus NIZO 3689 and Lactobacillus salivarius NIZO 3675). Individual probiotic strains carry the European Union qualified presumption of safety and all the study products (both probiotic and placebo) were prepared under good manufacturing process conditions. Each patient consumed sachets containing 5 g Ecologic® AAD or placebo twice daily for 2 weeks. Sachets had to be dissolved in lukewarm water, left for 10 min, stirred and thereafter ingested. Both probiotic and placebo were packaged in identical, numbered sachets. The placebo sachets, indistinguishable in colour, smell and taste also contained 5 % mineral mix and 15 % Raftilose® Synergy1 but contained maizestarch instead of the probiotic bacteria. In a pilot study with five healthy volunteers, the 2-week consumption of 1·5 g Raftilose® Synergy1 per day affected neither bifidobacterial communities (using PCR-denaturing gradient gel electrophoresis (DGGE) and most probable number-PCR) nor faecal lactic acid bacteria and lactobacilli counts (CJM Koning, D Jonkers, E Stobberingh, H Smid and R Stockbrügger, unpublished results).
Sample processing
Faecal samples were brought to the laboratory within 12 h after defecation and divided into three portions: (1) 10 g were centrifuged at 47 000 g for 2 h at 4 °C to obtain faecal water, which was frozen immediately in twofold at − 80°C for analysis of endotoxin concentrations and determination of pH; (2) 5 g were diluted (1:4) with peptone water (Oxoid CM9, Basingstoke, Hants, UK) supplemented with cysteine (2·1 mm) and glycerol (30 %) and used immediately for bacterial culture; (3) the remaining faecal sample (1–15 g) was frozen directly at − 20°C for molecular analysis.
DNA extraction
DNA was isolated from approximately 0·1 g of frozen faeces, using the FastDNA SPIN Kit for Soil (Qbiogene, Carlsbad, CA, USA) and a FastPrep Instrument (FP120, Savant Instruments, Farmingdale, NY, USA) following instructions of the manufacturer. Extracted DNA was checked by agarose gel (1·2 % (w/v)) electrophoresis in the presence of ethidium bromide.
PCR amplification, denaturing gradient gel electrophoresis analysis and quantitative PCR
Total bacterial profiles from all the samples were analysed by DGGE of 16S rRNA gene fragments amplified by PCR with primers 0968-f-GC and 1401-r as described previously(Reference Nubel, Engelen and Felske27, Reference Sousa, Pereira and Stams28). Amplicons generated by PCR were separated by DGGE in 8 % polyacrylamide gels using a DCode TM system (Bio-Rad Laboratories, Hercules, CA, USA) as described elsewhere(Reference Sousa, Pereira and Stams28, Reference Muyzer, de Waal and Uitterlinden29). For the separation of amplicons, gradients of 40–50 % were used (100 % denaturing solution was defined as 7 m-urea and 40 % formamide). Gels were stained with AgNO3(Reference Sanguinetti, Dias Neto and Simpson30). The DGGE gels were analysed using Bionumerics 4.0 (Applied Maths BVBA, Sint-Martens-Latem, Belgium). Following normalisation of gel images using markers loaded on either side and in the central lane of each gel, bands were defined for each sample using the bands searching algorithm within the program. Corresponding densitometric curves were used for a manual check of band definition, and bands with < 1 % of the total area of all bands were omitted from further analysis(Reference Konstantinov, Zhu and Williams31). The similarity of the DGGE profiles (expressed as similarity indices (SI)) with baseline (day 0; e.g. 0–7, 0–14, etc.) and between consecutive days (e.g. 0–7, 7–14, etc.) was determined by calculating a Dice correlation coefficient (band based) according to the principle of moving window correlation(Reference Possemiers, Verthe and Uyttendaele32).
Quantitative detection of total bacteria, lactobacilli and bifidobacteria was performed using an iCycler IQ real-time detection system associated with the iCycler optical system interface software version 2.3 (Bio-Rad, Veenendaal, The Netherlands). Total bacterial 16S rRNA gene copies were quantified with primers Bact-1369-f and Prok-1492-r(Reference Suzuki, Taylor and DeLong33), lactobacilli were quantified using primers LactoF and LactoR(Reference Byun, Nadkarni and Chhour34) and bifidobacteria by primers Bif 164-f and 662-r(Reference Satokari, Vaughan and Akkermans35) as described previously(Reference Fuentes, Egert and Jimenez-Valera36).
Bacteriological culture
Microbiological analysis of the faecal samples was determined as previously described(Reference Koning, Jonkers and Stobberingh37). Briefly, tenfold serial dilutions of the faecal dilution were made in physiological saline (0·85 %) with cysteine–HCl (0·05 %), and 40 μl of these dilutions were inoculated on agar plates to culture and count total (facultative) aerobic bacteria (blood agar, Oxoid CM 271), enterobacteria (methylthionium chloride agar, Oxoid CM69), enterococci (KF-streptococcus agar, Oxoid CM701), total (facultative) anaerobic bacteria (fastidious anaerobic agar, Laboratory M LabGo), Bacteroides spp. (Bile Esculin agar, Becton Dickinson 287920), lactobacilli (LAMVAB agar(Reference Hartemink, Domenech and Rombouts38)) and yeasts (Sabouraud GM+C agar, Becton Dickinson 254041). Escherichia coli and enterococci isolates were identified using standard biochemical tests(Reference Bruinsma, Hutchinson and van den Bogaard39) and stored at − 80°C for susceptibility testing.
Susceptibility testing
Antimicrobial susceptibilities (as minimal inhibitory concentration values) for the E. coli and enterococci isolates were determined using the microbroth dilution method with cation-adjusted Mueller–Hinton broth II (Becton, Dickinson and Company, Sparks, MD, USA) according to the Clinical and Laboratory Standards Institute (formerly the National Committee for Clinical Laboratory Standards) guidelines as described previously(Reference Nys, Okeke and Kariuki40). Microtitre plates containing freeze-dried antibiotics were obtained from MCS Diagnostics B.V. (Swalmen, The Netherlands) after quality testing by the manufacturer and a guaranteed shelf-life of about 1 year. The minimal inhibitory concentration was defined as the lowest concentration showing no growth in the microtitre plates after 18 h of incubation at 37°C. E. coli ATCC 35 218 and ATCC 25 922 and Escherichia faecalis ATCC 29 212 were used as quality control reference strains.
Endotoxin and pH
The endotoxin (i.e. lipopolysaccharide) concentration was determined in faecal water using the Limulus amoebocyte lysate endochrome technique (Endosafe, end-point chromogenic analysis endochrome test kit; Charles River, Kent, UK), according to the manufacturer's specifications under pyrogen-free conditions. Pyrogen-free water was used to dilute the faecal samples, the test solutions, and was used as a negative control. The detection range of the assay was 0·015–0·12 endotoxin units/ml (9 endotoxin units/ng). Concentration of faecal endotoxin was expressed as nanogram of endotoxin per ml of faecal water. The pH of faecal water was determined using a PHM standard pH meter with a PHC3006 electrode (Radiometer Nederland B.V., Zoetermeer, The Netherlands).
Statistics
The treatment allocation was concealed to all investigators and volunteers, until the study had been completed and all the analyses had been performed.
The primary outcome of the present study was to assess the restoration of the dominant intestinal microbiota after antibiotic treatment in probiotic- and placebo-treated patients by PCR-DGGE. Based on data from our previous study in healthy volunteers(Reference Koning, Jonkers and Stobberingh10), it was estimated that fourteen volunteers per treatment group would provide a 80 % power to detect a 20 % increase in SI after disturbance of antibiotic intake, assuming a variance of 18 % and a two-sided significance level of 0·05.
Secondary outcomes were to assess changes in probiotic- and placebo-treated patients during and after antibiotic intake in (1) specific bacterial subgroups, using both culture-dependent and molecular based techniques; (2) bowel habits; (3) the incidence of antibiotic resistance; (4) endotoxin concentration and pH. In addition, the disturbance of the dominant intestinal microbiota in the subjects who develop AAD compared with those who did not was assessed.
Statistical evaluation of DGGE band richness, quantitative PCR, culture, endotoxin concentration and pH was carried out using linear mixed model analysis as described previously(Reference Koning, Jonkers and Stobberingh10, Reference Koning, Jonkers and Stobberingh37).
For all other data the nonparametric Mann–Whitney U test was used for two-group comparisons of independent ordinal and interval values, while the nonparametric Wilcoxon signed-ranked test was used for comparison of related ordinal and interval values. If the data were normally distributed, the Student's t-test was used. For dichotomous variables, Pearson's χ2-test was applied to test for differences between groups. All the tests were conducted using SPSS version 11.0 (SPSS, Inc., Chicago, IL, USA) and a P value below 0·05 was considered statistically significant.
Results
Subjects
From September 2005 to December 2006, forty-five patients were enrolled of which thirty patients completed the study; seventeen in the probiotic group and thirteen in the placebo group. Eleven patients (73 %) discontinued the study in the first week, three patients (20 %) in the second week and one patient (7 %) thereafter. The most common reasons for discontinuation included ‘not being able or willing to collect faeces’; ‘perception of the study load as being too demanding considering the current medical condition’; ‘not liking the taste of the study product’. Baseline demographics and clinical characteristics were comparable for both the groups (Table 1). The antibiotic treatments prescribed for the exacerbation were mostly doxycycline and amoxycillin/clavulanic acid and were also comparable for both the groups. The mean frequency of antibiotic use was 3·1 (sd 2·4) treatments in the year before inclusion.
Table 1 Patient characteristics*
(Mean values and standard deviations)
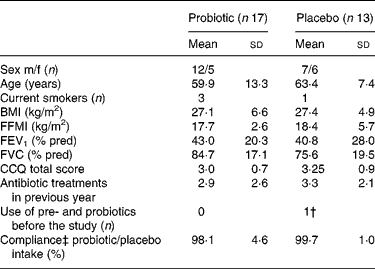
m/f, Male/female; FFMI, fat free mass index; FEV1, forced expiratory volume in 1 s; FVC, forced vital capacity; CCQ, clinical chronic obstructive pulmonary disease questionnaire; dd, developmentally disabled.
* There was no significant difference between the probiotic and the placebo groups (P>0·05).
† Yakult 1 dd.
‡ Compliance is self-reported.
Eight patients in the probiotic group and five patients in the placebo group (P = 0·72) changed their dietary habits during the study period, mostly based on medical indication (energy restriction (n 10), supplementation with nutrient drinks (n 2), loss of appetite (n 1)). Apart from the exclusion criteria, patients in both the groups used diverse and extensive medication mostly belonging to the groups of analgesia, antidepressants, antihypertensives, diuretics, sympathicomimetics, xanthines and parasympathicolytics.
There were no reported adverse events related to the study product. Compliance was assessed by the self-reported number of study product (sachets) that was consumed.
Bowel habits
The mean defecation frequency (probiotic, 1·4 (sem 0·4), 2·0 (sem 0·4), 1·6 (sem 0·3); placebo, 1·1 (sem 0·1), 1·4 (sem 0·2), 1·6 (sem 0·3)) and consistency (probiotic, 3·9 (sem 0·4), 4·5 (sem 0·3), 4·6 (sem 0·2); placebo 3·5 (sem 0·4), 4·2 (sem 0·4), 4·0 (sem 0·4)) before antibiotic and probiotic intake (day 0, i.e. baseline), during the antibiotic/probiotic period (days 1–7) and during the probiotic only period (days 8–14), respectively, did not differ significantly between the probiotic and the placebo groups. In the probiotic group, the defecation frequency during antibiotic intake was significantly higher (2·0 (sem 0·4)) compared with baseline (1·4 (sem 0·4); P < 0·05). During and 1 week after antibiotic intake (days 1–14), diarrhoea-like bowel movements were reported frequently in both the probiotic group (77 %) and the placebo group (70 %; P>0·05).
PCR-denaturing gradient gel electrophoresis analysis of the dominant faecal microbiota
DGGE analysis of the dominant faecal microbiota showed high inter-individual variation in total bacterial profiles between patients and a mean number of 15·4 bands per profile (i.e. band richness) at the start of the study (day 0). No difference in band richness between the probiotic and the placebo groups or over time within both the groups was found (Fig. 1).

Fig. 1 Total bacterial band richness (mean (sem) no. of bands) obtained from denaturing gradient gel electrophoresis profiles of probiotic- (▧) and placebo (![]() )-treated patients before, during (days 1–7) and after antibiotic intake.
)-treated patients before, during (days 1–7) and after antibiotic intake.
SI were high and remained stable during and after antibiotic treatment (Table 2). No effect of probiotic intake was observed. At day 63, lower SI (compared with baseline) were found in both the groups, which were only significant in the probiotic group. Moreover, no association between the SI of the total bacterial profiles during amoxycillin and probiotic intakes and the occurrence of diarrhoea-like bowel movements was observed (data not shown).
Table 2 Similarity indices (SI) of the total bacterial profiles in percentages in the probiotic group v. the placebo group compared with baseline (t=0) and between consecutive intervals
(Median and range values)
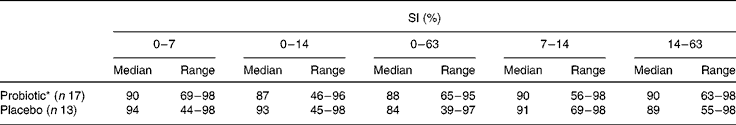
* Within group decrease 0–63 v. 0–7, P < 0·05.
Quantitative PCR
No differences in the 16S rRNA gene copy numbers of total bacteria, lactobacilli or bifidobacteria were observed between the probiotic and the placebo groups. Within both the groups, a significant decrease of bifidobacteria was found during antibiotic intake, which increased again after cessation (Fig. 2). During probiotic intake (day 14), a small increase in total bacteria was observed in the probiotic group, which significantly decreased again after cessation. Moreover, a significant increase in lactobacilli was observed in the probiotic group (Fig. 2). For all the bacterial species studied, values on day 63 did not differ significantly from day 0 apart from a decrease in bifidobacteria in the probiotic group and a decrease in total bacteria in the placebo group (Fig. 2). To adjust for possible differences in faecal consistency, data were also analysed as 16S rRNA gene copies per mg faecal protein with similar results compared with the unadjusted data (data not shown).

Fig. 2 Quantitative PCR results expressed as mean (sem) log copies DNA/g faeces obtained from faeces of probiotic- (▧) and placebo (![]() )-treated patients before, during (days 1–7) and after antibiotic intake: *P < 0·05.
)-treated patients before, during (days 1–7) and after antibiotic intake: *P < 0·05.
Bacteriological culture
No differences in either aerobic or anaerobic cultured-bacterial species were observed between the probiotic and the placebo groups (Table 3). However, group-specific differences were observed over time. During ‘the probiotic only period (14 d)’, the mean number of faecal enterococci increased significantly within the probiotic group, returning to pre-treatment level 7 weeks after cessation (P < 0·05; Table 3). Moreover, within the probiotic group, a significant decrease was found in enterobacteria (day 7 v. day 0), and significant increases were observed over time in total aerobes (days 7 and 14 v. days 0 and 63), Bacteroides spp. (day 14 v. days 0 and 63) and lactobacilli (days 14 and 63 v. day 7) (P < 0·05). Within both the groups, a significant increase in yeast was found during antibiotic intake (day 7; Table 3).
Table 3 Numbers of bacteria cultured expressed as log colony-forming units/g faeces
(Mean values with their standard errors)

* Within group increase t = 7/14 v. t = 0/63, P < 0·05.
† Within group decrease t = 7 v. t = 0, P < 0·05.
‡ Within group increase t = 14 v. t = 0/7/63, P < 0·05.
§ Within group increase t = 14 v. t = 0/63, P < 0·05.
∥ Within group increase t = 14/63 v. t = 7, P < 0·05.
¶ Within group increase t = 7 v. t = 0, P < 0·05.
** Within group increase t = 7 v. t = 0/63, P < 0·05.
For all the bacterial species, in both the groups, bacterial counts on day 63 did not differ significantly from day 0.
Antimicrobial susceptibility
E. coli isolates were isolated in 47–82 % (probiotic) and 54–69 % (placebo) of the faecal samples and enterococci were isolated in 77–100 % (probiotic) and 92 % (placebo) of the faecal samples from the patients at the different points of time. No significant differences in antibiotic susceptibility of E. coli and enterococci were observed.
Endotoxin and pH
Mean endotoxin concentrations, expressed as log ng/ml faecal water, did not differ significantly between groups and within both groups during the study period (probiotic, 2·5 (sem 0·1), 2·6 (sem 0·08), 2·5 (sem 0·07) and 2·4 (sem 0·09); placebo, 2·5 (sem 0·06), 2·5 (sem 0·09), 2·5 (sem 0·09), 2·5 (sem 0·01) at t = 0, 7, 14 and 63, respectively). Also, no significant correlation between the number of enterobacteria cultured and endotoxin concentration was found.
The pH of faecal water ranged from 6·3 to 6·7 and did not differ between groups and within both groups during the study period.
Discussion
The present double-blind, placebo-controlled, randomised study demonstrated that the intake of a multispecies probiotic in this COPD population with a history of frequent antibiotic use had no effect on the composition of the dominant faecal microbiota. Moreover, in contrast to previous findings in healthy volunteers, no effect of antibiotic intake on the composition of the dominant faecal microbiota could be found and no difference in SI was observed between the group of patients who developed diarrhoea-like bowel movements and those who did not. Looking at specific bacterial subgroups, using both molecular and culture-dependent techniques, a significant but small antibiotic effect was observed in both the groups, i.e. an increase in yeasts and a decrease in bifidobacteria. No differences were observed when comparing the probiotic group with the placebo group. However, changes in specific bacterial groups over time were seen in the probiotic group only, suggesting a modest probiotic effect.
In healthy human subjects, the faecal microbiota is host specific and relatively stable over time (SI>80 %)(Reference Vanhoutte, Huys and De brandt5, Reference Zoetendal, Akkermans and De Vos6). It is well documented that despite methodological differences, antibiotic treatment, especially amoxycillin, cephalosporins, clindamycin or broad-spectrum antibiotics, has a marked effect on the composition of the intestinal microbiota(Reference Dethlefsen, Huse and Sogin8, Reference De La Cochetiere, Durand and Lepage41, Reference Sullivan, Edlund and Nord42), which may result in diarrhoea. In line with these findings, we have previously shown that amoxycillin intake strongly affected the composition of the dominant faecal microbiota in healthy volunteers, indicated by a low mean SI ( < 52 %) and a decrease in bacterial richness (i.e. number of bands). We also showed that a lower stability of the dominant intestinal microbiota (i.e. more disruption) is related to a higher risk of developing AAD(Reference Koning, Jonkers and Stobberingh10). Remarkably, using the same methods, in the COPD population mean SI were high (>90 %) and both the SI and the band richness remained stable during and after antibiotic intake. Moreover, no difference in SI (i.e. disruption) was observed in the group of patients, who developed diarrhoea-like bowel movements compared with those who did not. This unexpected lack of effect on the microbiota could be attributed to the frequent antibiotic use in the past by this population. Recently, it has been shown that antibiotic intake can cause medium and long-term disturbances in (specific) bacterial populations(Reference Jernberg, Lofmark and Edlund9, Reference Koning, Jonkers and Stobberingh10). Moreover, as antibiotics only affect sensitive strains, extensive antibiotic use could have caused a prolonged narrowing of the diversity into a dominant faecal microbiota consisting of microbial populations insensitive to the given antibiotics. This assumption is further supported by the fact that bacterial band richness in COPD patients at the start of the study was relatively low, i.e. being at the same level as in healthy volunteers during amoxycillin intake, in whom band richness was significantly reduced(Reference Koning, Jonkers and Stobberingh10). It is known that the bacterial diversity decreases with age(Reference Woodmansey43), and we readily acknowledge that the mean age of the patients in the present study was significantly higher than that of the healthy volunteers in the previous study. However, this cannot account for the fact that no antibiotic effect on bacterial diversity was observed in the COPD patients. In fact, this finding was corroborated by a similar trend in endotoxin concentrations. Previously, antibiotics have been shown to increase the bioavailability of endotoxin originating from gram-negative bacteria(Reference Goris, de Boer and van der Waaij44–Reference Hurley47). However, in our study, we found that endotoxin concentrations remained stable during and after antibiotic intake and were even slightly higher than the levels found in healthy volunteers during amoxycillin intake, where an increase was found(Reference Koning, Jonkers and Stobberingh37).
In addition to qualitative data on microbiota composition obtained by DGGE analysis of 16S rRNA gene fragments, quantitative analysis of specific bacterial subgroups was performed by culture and quantitative PCR.
With culture, apart from a small but clinically insignificant rise in yeasts, no changes were observed, that could be attributed to antibiotic intake. Moreover, with quantitative PCR, no effect of antibiotic intake was found on total bacteria, which is in line with the DGGE results. This further supports the lack of observed antibiotic effect. However, the number of bifidobacteria was significantly affected in both the groups during antibiotic intake corresponding with the reported high susceptibility of bifidobacteria to broad-spectrum antibiotics(Reference Ammor, Florez and Mayo48–Reference Woodmansey, McMurdo and Macfarlane51).
Various effects over time were observed in the probiotic group only, which can be attributed to probiotic intake. The increase in faecal enterococci and lactobacilli during probiotic intake is in line with literature showing that the intake of a probiotic is able to transiently increase the faecal number of the ingested strains(Reference Goossens, Jonkers and Russel52–Reference Tannock, Munro and Harmsen54). We previously showed that the consumption of this multispecies probiotic, containing E. faecium, was associated with a significant increase in the concentration of faecal enterococci in the probiotic group and that the pulsed-field gel electrophoresis patterns of the enterococci cultured were similar to the orally administered E. faecium (Reference Koning, Jonkers and Stobberingh37). Apart from lactobacilli and the E. faecium, the multispecies probiotic also contained bifidobacteria. However, no increase in bifidobacteria could be observed, probably due to the fact that the administered daily amount (2 × 109 colony forming units) was too low in relation to the baseline value.
The multispecies product was well tolerated and no adverse events related to the product were reported. The majority of literature shows that the probiotics can significantly reduce the relative risk of developing AAD(Reference Cremonini, Di Caro and Nista19, Reference D'Souza, Rajkumar and Cooke20, Reference McFarland22). In contrast, in the present study, the intake of the multispecies probiotic did not result in a decrease in diarrhoea-like bowel movements. However, earlier we showed, in healthy volunteers, that the intake of the same multispecies probiotic had a restoring and stabilising impact on the dominant faecal microbiota after amoxycillin intake(Reference Koning, Jonkers and Stobberingh10). A comparable observation was made by Kajander et al. (Reference Kajander, Myllyluoma and Rajilic-Stojanovic55), when a multispecies probiotic was given to irritable bowel syndrome patients. In the present study, antibiotic intake did not affect the composition of the dominant microbiota, probably contributing to the lack of effect of probiotic intake on the occurrence of diarrhoea-like bowel movements in this COPD population. Furthermore, as the microbiota seems to be profoundly altered by the frequent prior antibiotic use, a much longer probiotic treatment might be required to restore such an effect.
We acknowledge that the present study has some limitations. Inherent to the nature and severity of the disease, diverse and extensive medication was used in this COPD population, and we cannot exclude that some medication had an impact on the composition of the microbiota. Nevertheless, patients had to be on stable medication for corticosteroids, gastric acid inhibitors and doxycycline maintenance therapy before and during the study to limit this effect. Although we clearly showed no effect of antibiotic treatment on the intestinal microbiota due to the heterogeneity of the population and the limited sample size, subtle changes in subgroups' analyses may have failed to be detected.
Currently, it is widely accepted that antibiotic treatment profoundly affects the intestinal microbiota. However, our findings show that the dominant faecal microbiota was not affected upon antibiotic treatment in COPD patients with a history of frequent antibiotic use. This prolonged antibiotic pressure seems to have caused a long-term imbalance of the microbiota with a shift toward populations resistant to antibiotics. Furthermore, the present study demonstrates that the short-term intake of a multispecies probiotic had a modest effect on the intestinal microbiota, albeit much smaller than previously observed for healthy adults. Nevertheless, this did not result in a restoration of the microbiota imbalance and as a result no reduction in diarrhoea-like bowel movements was found.
These results warrant further confirmation in studies with a higher number of patients with higher doses of probiotics and a longer administration period.
In future studies, it would be very interesting to determine to what extent these disturbances are permanent, what the effect of these disturbances is and if probiotic supplementation is able to either prevent or with prolonged use reverse such an imbalance.
Acknowledgements
The authors would like to thank Annie van de Kruijs (Centre for Integrated Rehabilitation of Organ failure, Horn, The Netherlands) for expert assistance with the enrolment of patients and collection of samples, and Wilma Akkermans-van Vliet and Ineke Heikamp-de Jong (Laboratory of Microbiology, Wageningen University, The Netherlands) for excellent assistance with the PCR-DGGE analyses. The present study was supported by SenterNovem, an agency of the Dutch Ministry of Economic Affairs (grant no. TSGE 1041). R. S. is the guarantor of the article. C. K. was the principal investigator and conducted the study. C. K., D. J. and H. S. were responsible for the analyses of the study. All the authors participated in the design, execution and data interpretation of the study and contributed significantly to the various drafts of the manuscript. All the authors approved the final draft submitted. F. R. is an advisor of Winclove Bio Industries B.V. C. K. is supported, in part, by the grant from SenterNovem and, in part, by Winclove Bio Industries B.V. All the other authors declare they have no conflict of interest.







