INTRODUCTION
Campylobacter causes an estimated 1·3 million illnesses in the USA each year [Reference Scallan1]. Antimicrobial resistance in this pathogen is a serious concern [2]. There are currently 26 Campylobacter species; Campylobacter jejuni accounts for the majority (85–95%) of human infections, followed by C. coli (5–10%), and less commonly C. upsaliensis, C. fetus, C. lari, and other rare species [Reference Fitzgerald3].
Speciation is important in understanding the epidemiology of infection because Campylobacter organisms and their reservoirs are diverse. The primary reservoirs are poultry, cattle, and pigs; however, Campylobacter has been isolated from numerous other animal species and the environment, and the prevalence of different Campylobacter species in animals differs. For example, C. coli has been isolated more often than C. jejuni from swine and C. jejuni more often than C. coli from chicken [4]. C. fetus is primarily associated with cattle and sheep [Reference Wagenaar5] but has also been identified in reptiles [Reference Gilbert6]. C. upsaliensis is frequently isolated from cats and dogs [Reference Acke7]. C. lari has been isolated from a variety of sources including wild birds, seawater, and shellfish [Reference Matsuda and Moore8]. Unpasteurized dairy products, poultry, and drinking water have been sources of Campylobacter outbreaks caused by various species [Reference Taylor9]. Antimicrobial susceptibility patterns differ by species, with more erythromycin and ciprofloxacin resistance in C. coli isolates [4]. Differences in demographics of and risk factors for infection with various Campylobacter species have been described in patients in Europe [Reference Gillespie10, Reference Bessède11].
Few clinical laboratories in the USA routinely perform speciation beyond C. jejuni. Species-level identification of non-C.jejuni is challenging using phenotypic methods and it does not affect clinical treatment nor aid in outbreak detection. In addition, isolation methods are biased toward recovery of C.jejuni. Thus, Campylobacter surveillance data in the USA are typically presented at the genus level. To our knowledge, no comprehensive description of Campylobacter cases at the species level in the USA has been published. We attempt to fill this gap by describing available species-level data from 10 Foodborne Diseases Active Surveillance Network (FoodNet) sites, and comparing characteristics of patients and illnesses by the species causing infection.
METHODS
FoodNet is the foodborne disease component of the Centers for Disease Control and Prevention (CDC)’s Emerging Infections Program, a collaborative project of CDC, 10 state health departments, the United States Department of Agriculture's Food Safety and Inspection Service, and the Food and Drug Administration. Since 2004, the FoodNet catchment area has included the entire states of Connecticut, Georgia, Maryland, Minnesota, New Mexico, Oregon, and Tennessee and selected counties in California, Colorado, and New York. FoodNet conducts surveillance for human Campylobacter infections by routinely contacting clinical laboratories serving the catchment area to compile reports of all culture-confirmed infections. Staff collect patient demographics, hospitalization status, outcome, and specimen information from physicians, laboratory records, or through a patient interview in all sites. Information on international travel in the 7 days before illness onset is recorded from an interview or patient chart when available, and patients with such a history are considered to have travel-associated infections.
In some sites, isolates are sent from clinical laboratories to state public health laboratories (SPHLs) for confirmation and speciation. Between 2010 and 2015, Maryland, Minnesota, and New Mexico required all isolates to be submitted, and New York required submission of isolates from 15 of 34 FoodNet counties; routine submission was requested but not required in Georgia and Tennessee, and routine submission was neither requested nor required in California, Colorado, Connecticut, and Oregon. In addition, all sites forward some isolates to CDC for speciation and antimicrobial susceptibility testing as part of the National Antimicrobial Resistance Monitoring (NARMS) Program. Seven states selected Campylobacter isolates to forward based on a sampling scheme applied to isolates received at SPHLs: Georgia, Maryland, and New York (one forwarded for every two received), Minnesota (1:5), New Mexico (1:3), Oregon and Tennessee (all). In California, Colorado, and Connecticut, isolates are sampled from those received at the SPHL from one participating reference laboratory in each state. Because not all states require isolates to be submitted to the SPHL, the number of isolates sent to CDC is not consistently proportional to the total number of Campylobacter infections reported to FoodNet.
Identification of C. jejuni was performed by the typical colony and Gram stain morphology, catalase, oxidase, growth at 42 °C, and the hippurate hydrolysis test. Hippurate-positive isolates were identified as C. jejuni. Isolates that were negative for hippurate hydrolysis were further tested with indoxyl acetate. Hippurate-negative isolates were also further characterized by polymerase chain reaction assays with species-specific targets for C. jejuni (mapA or hipO gene), C. coli (ceuE gene or glyA gene), C. fetus (sapD gene), or other species-specific primers [Reference Linton12]. C. jejuni and C. coli isolates were tested for antimicrobial susceptibility using standard NARMS methods [13]. Only resistance to quinolones (ciprofloxacin or nalidixic acid) and macrolides (azithromycin or erythromycin) is included in our analysis because these are the two most important classes for treatment.
We analyzed data from cases of Campylobacter infection reported to FoodNet from 2010 through 2015 that had species results available from a SPHL or CDC. Antimicrobial susceptibility information was available for 2010–2015 from a subset of C. jejuni and C. coli isolates. We performed statistical analyses using SAS version 9.3 (SAS Institute, Cary, NC, USA). Incidence rates were calculated using US census data. C. coli, C. lari, C. fetus, and C. upsaliensis were individually compared with C. jejuni by age (median and in 5-year categories), sex, race, ethnicity, clinical symptoms, hospitalization, outcome, specimen source, international travel, season, and residence in a metro area. Fisher's exact tests were used to compare proportions. The criterion for significance was a 95% confidence interval (CI) excluding 1·0. All variables that showed significant associations were included in species-specific multivariable logistic regression models. Final models were developed using the stepwise option in Proc Logistic SAS Version 9.3. (SAS Institute, Cary, NC, USA). Seasons were defined as follows: winter (December–February), spring (March–May), summer (June–August), and autumn (September–November). FoodNet sites were grouped by US Census Region as follows: northeast (CT, NY); midwest (MN); south (GA, MD, TN); mountain (CO, NM); and Pacific (CA, OR). FoodNet counties were classified as urban, suburban, or rural based on USDA's Economic Research Service 2013 Rural–Urban Continuum Codes [14].
RESULTS
We identified 39 345 culture-confirmed Campylobacter infections in FoodNet sites from 2010 through 2015. We obtained species results for 16 549 (42%) of the isolates; 6971 (42%) were speciated at CDC, and 9578 (58%) were speciated at SPHLs. Forty-four percent of the results from SPHLs came from Minnesota. Of 16 549 isolates with species information, 14 672 (89%) were C. jejuni, 1404 (8%) were C. coli, 333 (2%) were C. upsaliensis, and 140 (1%) were other species, including C. lari (98), C. fetus (35), Campylobacter hyointestinalis (five), Campylobacter curvus (one), and Campylobacter helveticus (one). The percentage of non-C. jejuni species was relatively stable at 10% (259/2673) in 2010 and 12% (336/2784) in 2015.
Patient age and sex distributions varied across Campylobacter species (Fig. 1 and Table 1). Patients with C. coli infection tended to be older than C. jejuni patients but had a similar sex distribution, with about 56% of infections in males. C. upsaliensis patients tended to be younger – the proportion of patients <5 years old was higher than for any other species – and a higher percentage (58%) were female. Although few patients with C. fetus and C. lari infection were reported, their age and sex distribution showed patterns similar to but more pronounced than those of C. coli, with even higher median ages (50 and 57 years) and percentages of males (83%, 63%), respectively. The majority of persons with Campylobacter infection identified as white and non-Hispanic, but there were some racial and ethnic differences between patients infected with different species. Thirteen percent of persons with C. fetus cases were Black compared with 8% or less for other species. Nine percent of C. fetus and 6% of C. coli patients were Asian, compared with 5% or less for other species. Sixteen percent of C. upsaliensis patients were Hispanic, compared with 12% or less for other species.
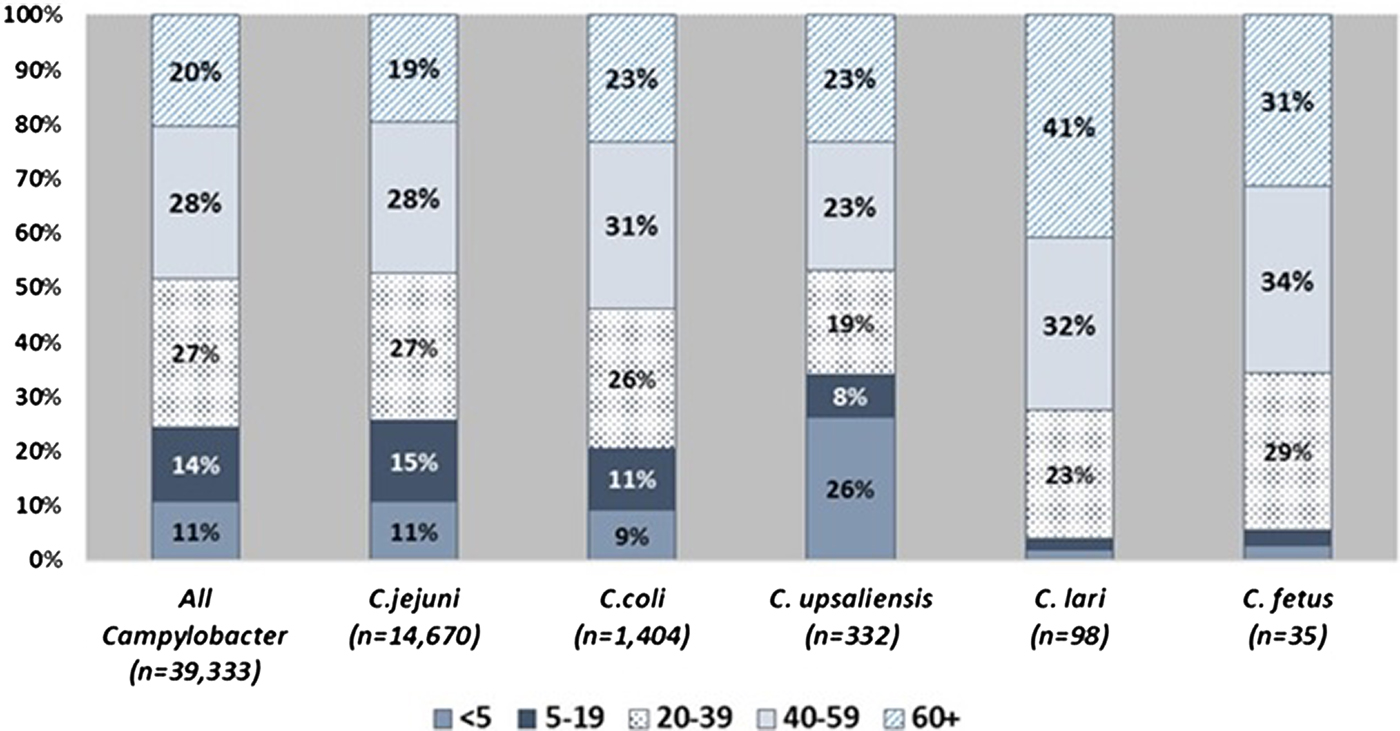
Fig. 1. Distribution of patients with Campylobacter infection, by age group and species – FoodNet, 2010–2015.
Table 1. Characteristics of Campylobacter patients, by five most common species – FoodNet, 2010–2015
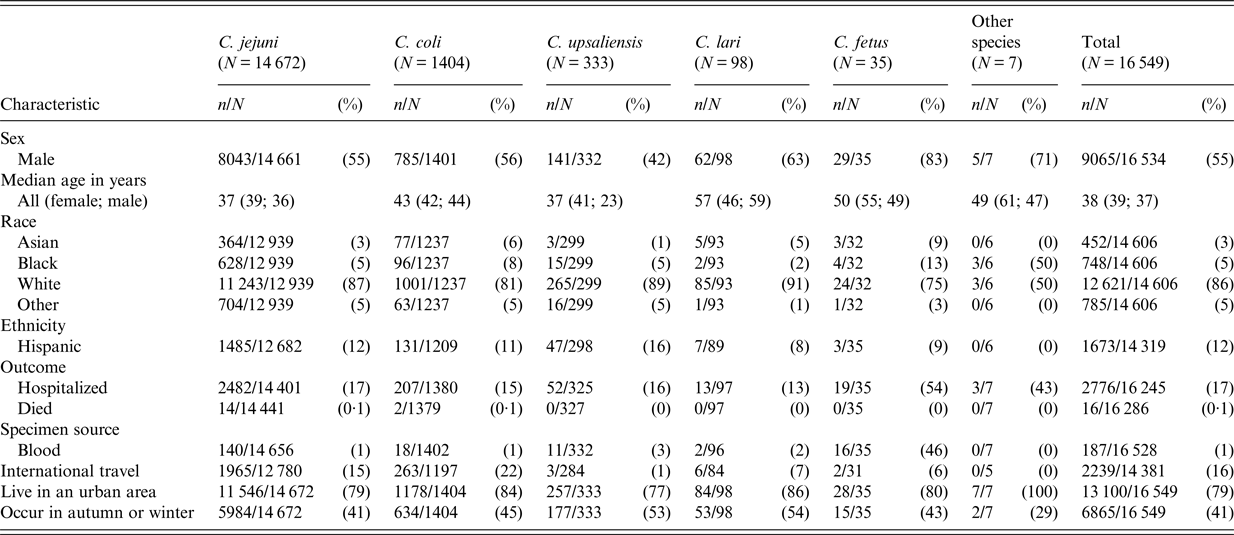
Specimen source and patient outcome also varied by species, especially for C. fetus. The proportion of isolates from blood was highest (46%) for C. fetus infections, and over half of C. fetus patients were hospitalized, compared with ⩽3% bloodstream isolations and ⩽17% hospitalization for most other species.
C. jejuni and C. coli infections had similar regional distributions. A higher percentage of C. upsaliensis cases occurred in the Pacific region, C. lari cases in the south, and C. fetus cases in the midwest (Fig. 2). C. jejuni incidence rates were highest in rural areas for persons younger than 60 years and in suburban areas for persons 60 years and older (Fig. 3). C. coli rates were highest in rural areas for children <5 years, in suburban areas for persons aged 5–39 years, and in urban areas for persons 40 years and older. C. upsaliensis rates were highest in rural areas for children <5 years and persons over 60 years. These patterns persisted after excluding travel-associated cases.
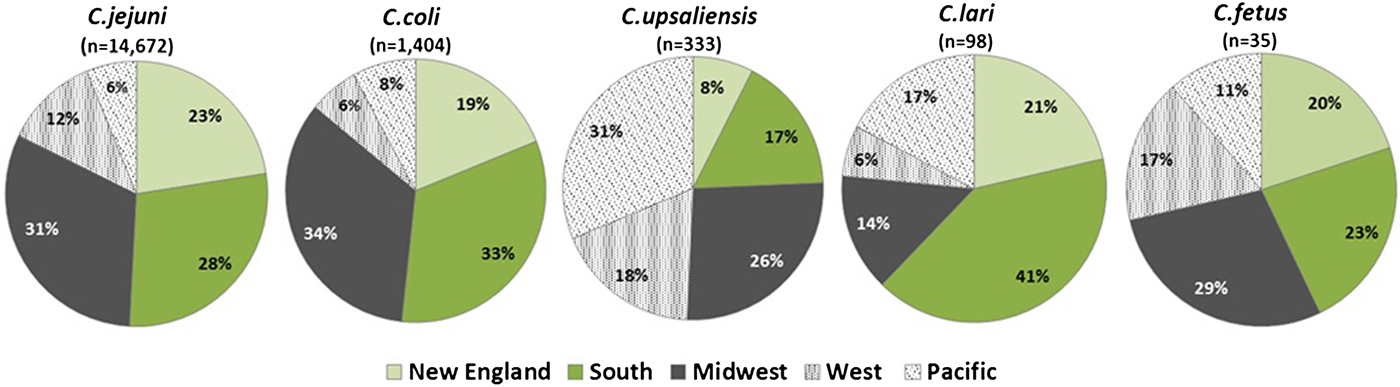
Fig. 2. Distribution of Campylobacter species by geographic region – FoodNet, 2010–2015.
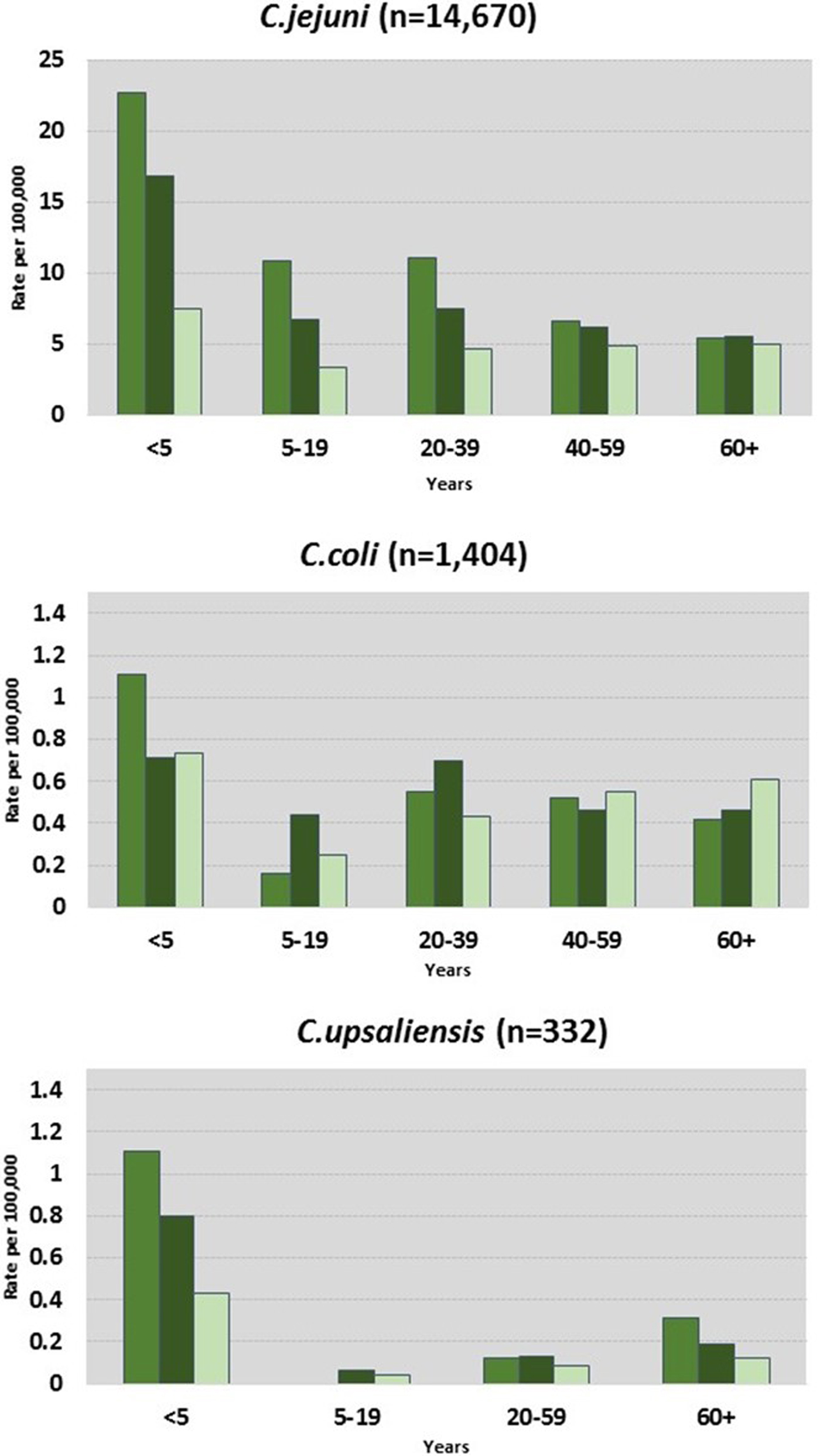
Fig. 3. Campylobacter infections by species, location, and age group – FoodNet, 2010–2015.
C. jejuni and C. coli infections exhibited strong summer peaks; the peak in C. coli cases occurred slightly later than C. jejuni. C. upsaliensis and C. lari infections predominated in autumn and winter months (Fig. 4). International travel in the 7 days before illness onset was reported by 16% of persons with Campylobacter infection, most commonly among those with C. coli (22%) and rarely among those with C. upsaliensis (1%) infections (Table 1). Travel-associated Campylobacter infections showed little seasonality with the exception of a slight winter peak among C. coli cases (Fig. 4).
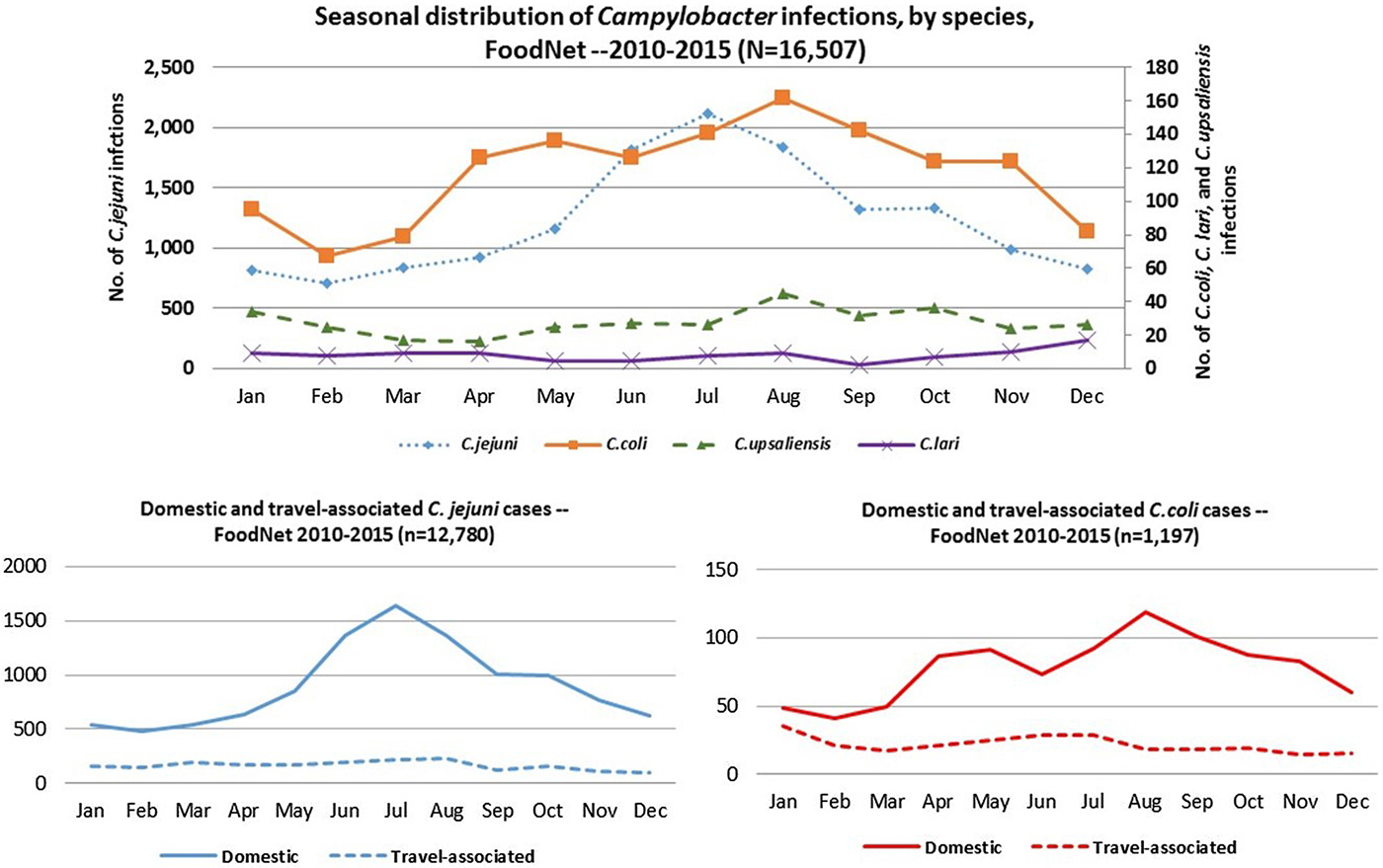
Fig. 4. Seasonal distribution of patients with Campylobacter infection by species, overall and by international travel history – FoodNet, 2010–2015.
Data from patients with infections of the more common Campylobacter species were compared with data from C. jejuni patients in multivariate models (Table 2). C. coli patients were more likely than C. jejuni patients to be older than 40 years, be Asian, be Black, have an infection in fall or winter, live in an urban area, and report international travel in the 7 days before infection; and less likely to be hospitalized. C. upsaliensis patients were more likely than C. jejuni patients to be female, be Hispanic, have an isolate from blood and infection in autumn or winter, and less likely to report international travel. C. lari patients were more likely than C. jejuni patients to be older than 40 years and have an infection in autumn or winter. C. fetus cases were more likely than C. jejuni cases to be male, hospitalized, and have an isolate from the blood.
Table 2. Multivariate analysis of selected Campylobacter species compared with Campylobacter jejuni, FoodNet, 2010–2015

n/s, not statistically significant.
Antimicrobial susceptibility information was available for 6010 C. jejuni and 665 C. coli isolates. Resistance to quinolones (35% vs. 24%) and macrolides (11% vs. 3%) was higher among C. coli isolates than C. jejuni isolates. Resistance among C. jejuni isolates was higher in the metro (27%) than in suburban and rural (15%) areas (P < 0·0001). Among patients with C. jejuni infection, age >20 years (OR = 1·4; 95% CI 1·1–1·8), Asian race (OR = 2·3; 95% CI 1·4–3·9), and international travel in the 7 days before infection (OR = 12·5; 95% CI 10·0–15·7) was associated with any resistance. Among C. coli patients, international travel in the 7 days before infection (OR = 12; 95% CI 6·4–22·7) was associated with any resistance. Among travelers with either species, the highest percentage of resistant infections was found among persons who visited Asia (18/171 (91%)) or South America (17/83 (83%)).
DISCUSSION
Public health activities to control Campylobacter infections in the USA generally focus at the genus level; speciation of isolates is relatively unusual. Our analysis showed significant demographic and clinical differences between patients infected with different species highlighting the value of examining Campylobacter at the species level. The high burden of Campylobacter infection, its associated sequelae such as Guillain–Barré syndrome, and the potential for treatment failure due to antimicrobial resistance emphasize the urgency of improved control.
C. jejuni is the most common species and provides a good comparison group against which to examine characteristics of other species. In our data, patients with C. jejuni infection were predominately White and non-Hispanic, and a little over half were male. Less than a quarter of patients were hospitalized, and few had invasive infections. Rates of infection were highest in rural areas, particularly among children <5 years old, an association that has been well documented in other countries [Reference Green, Krause and Wylie15, Reference Ethelberg16]. C. jejuni infections overall showed a strong summer seasonality consistent with patterns described for other temperate countries [Reference Nylen17]. It is well established that poultry is the primary risk factor for sporadic infections [Reference Doorduyn18, Reference Friedman19]. Although poultry is eaten year-round, the seasonality of infections may be explained by studies that have shown that the prevalence of Campylobacter in broiler flocks and poultry at retail varies by season and is highest in warmer months [Reference Boysen, Vigre and Rosenquist20, Reference Willis and Murray21]. Exposure to other domestic risk factors common in summer months such as animal contact, raw milk consumption, swimming, eating barbeque-prepared meals, and drinking water from untreated sources likely also influence the summer peak [Reference Friedman19, Reference Schonberg-Norio22].
C. coli is the second most commonly identified species in the USA. Compared with C. jejuni, persons infected with C. coli were more often older than 40 years, Asian, or Black. Nearly a quarter of patients with C. coli reported international travel. Many of these differences are consistent with previous studies [Reference Gillespie10, Reference Bessède11, Reference Roux23], but the association with Black race, to our knowledge, has not been previously reported. Mechanisms behind these differences are not known but could reflect occupational or environmental exposures, cultural practices, dietary preferences, or genetic predisposition. The use of proton pump inhibitors has been associated with Campylobacter infection [Reference Doorduyn18, Reference Neal24], and differential resistance to acid between the two species has been hypothesized to play a role in age differences [Reference Roux23]. C. coli infection has been associated with the consumption of pate and meat pies [Reference Gillespie10], bottled water [Reference Gillespie10], undercooked eggs [Reference Sopwith25], swimming [Reference Doorduyn18], and contaminated surface water [Reference Zeigler26]. International travel might contribute to the higher rate of C. coli infections in winter months.
C. upsaliensis is the third most commonly identified species in the USA. Compared with C. jejuni, patients with C. upsaliensis infection were more often female and Hispanic. Of all the species we examined, C. upsaliensis infections were least likely to be travel-associated. Female gender has been associated with infection with specific C. coli and C. jejuni strain types found in poultry [Reference Bessell27], possibly because women are more likely than men to prepare food, but it is unclear whether this explanation extends to other species. While C. upsaliensis has been isolated from foods such as ground beef [Reference Trokhymchuk28], the infection has more often been associated with exposure to domestic pets such as dogs and cats. The association with Hispanic ethnicity is unexplained. It could reflect food preferences or be linked to cultural practices and should be further explored, but we do not think it is related to exposures abroad, because the travel-associated proportion is so low. Our finding that C. upsaliensis infections occur more often in autumn or winter months is intriguing but should be interpreted with caution given the small sample size.
We identified a small number of infections with other Campylobacter species. Patients with C. fetus and C. lari infections were predominately male and tended to be older than those with C. jejuni. Older age and male predominance has been previously documented for C. fetus [Reference Wagenaar5, Reference Gazaigne29] but is not well understood. It is possible that occupational exposure to cattle and sheep plays a role as many sporadic C. fetus cases reported in the literature have occurred among farmers or abattoir workers [Reference Wagenaar5]. Patients with C. fetus infection were much more likely to have bloodstream infections and to be hospitalized than patients infected with any other species. Host factors, pathogen factors, or both may explain this characteristic. C. lari is associated with gastrointestinal infections and bacteremia especially in persons who are immunocompromised. It has been isolated from multiple environmental sources, including surface water, wild birds, and domestic animals [Reference Matsuda and Moore8], and contaminated water has been associated with infection [Reference Broczyk30].
Consistent with previous studies [Reference Van Hees31, Reference Kassenborg32], we found international travel to be a risk factor for antimicrobial-resistant infections, particularly among C. coli patients, and a high percentage of resistant infections occurred among travelers to Asia and South America. Resistance to ciprofloxacin is known to be common in these regions and appears to be increasing [Reference Pollett33, Reference Post34]. Ciprofloxacin resistance among travelers is a concern because ciprofloxacin is a common treatment for diarrhea in returned travelers [Reference Steffen, Hill and DuPont35], although a macrolide is the drug of choice for Campylobacter infections [Reference Allos, Blaser, Mandell, Bennett and Dolin36]. We also found that a higher percentage of antimicrobial-resistant infections occurred in urban areas. This finding is consistent with reports from the Netherlands; they hypothesized consumption of ready-to-eat foods may be higher in urban areas [Reference Van Hees31]. It is also possible that persons in urban areas have more severe illness or co-morbidities. Interventions to combat resistance in the USA should focus on targeting domestic food sources, enhancing awareness among healthcare providers and travelers about the increased risk of resistant infections associated with international travel, and promoting appropriate antimicrobial stewardship.
Many factors affect the availability of Campylobacter species data in the USA and these should be taken into account when interpreting our findings. Although infection has been reportable in most states, it did not become a nationally notifiable infection until 2015 [37]. Only 22 (44%) states request or require positive specimens to be sent to SPHL for confirmation and speciation [38], and there are currently no national guidelines for the isolation and speciation of Campylobacter. Only 31% of 403 laboratories serving the FoodNet catchment area routinely speciate Campylobacter [Reference Hurd39]. Thus, the process leading to the inclusion of cases in our dataset is complex, and the proportion of cases due to each species is not generalizable to all Campylobacter infection. Nonetheless, the differences we observe between species are unlikely to be due to selection bias, as these pre-speciation biases would be expected to affect all species similarly.
Many laboratories rely on isolation methods and selective media optimized for recovery of C. jejuni from the stool, decreasing the likelihood of recovering many other Campylobacter species. Selective media for isolation of specific non-jejuni species either has not been developed or is not widely available. Isolation methods, such as filtration, that would increase recovery of non-jejuni species are labor-intensive and few laboratories perform them. Traditional phenotypic identification methods [38] are widely performed and accurately identify hippurate-positive C. jejuni. However, these methods do not accurately identify hippurate-negative Campylobacter species or hippurate-negative C. jejuni. A method for species-level identification of Campylobacter using matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF) has been described [Reference Mandrell40] and is becoming more widely used by clinical laboratories and SPHLs. DNA-based molecular methods can enhance species identification but can be expensive to implement. Since 2010, there has been an increase in the proportion of infections caused by species other than C. jejuni. This could reflect a real change in sources of human Campylobacter infections. Alternatively, it could be due to increased awareness of the non-jejuni species and improved ability to accurately identify these species in the state health departments after the implementation of CDC's quality assurance program for the identification of Campylobacter in 2008 and follow-up training made available to the SPHLs.
The use of culture-independent diagnostic tests (CIDTs) for identification of Campylobacter in FoodNet sites is increasing [Reference Iwamoto41]. Decreasing availability of Campylobacter isolates has important implications for public health surveillance. While CIDTs are quicker and easier to perform, the most commonly used tests at present are limited in the Campylobacter species detected. They do not differentiate between Campylobacter species, nor do they yield an isolate for speciation or antimicrobial susceptibility testing. A strategy for obtaining species information, such as the reflex culture of specimens with a positive CIDT result, reflex culture if medically indicated for patient care, or collection of isolates from sentinel laboratories, is needed to improve understanding of species-related differences and inform source attribution and risk factor studies. Advanced molecular detection methods, such as whole genome sequencing, will greatly facilitate identification and classification of strains and comparison of strains from human, food, animal, and environmental sources, but these methods rely on the availability of isolates to test. Coupled with standardized case exposure data, these approaches offer promise for a clearer understanding of the epidemiology of Campylobacter species in the USA, which in turn can inform prevention strategies.
ACKNOWLEDGEMENTS
This work was supported by the Centers for Disease Control and Prevention (CDC-RFA-CK17-1701 – Emerging Infections Program).
DECLARATION OF INTEREST
None.









