Introduction
Salmonella Typhimurium outbreaks, UK
Non-typhoidal salmonellosis is caused by the enteric pathogen Salmonella enterica, a species that includes over 2,600 different serovars [Reference Guibourdenche1]. In the United Kingdom (UK), over 8,000 cases of salmonellosis are reported annually [2], with the most commonly reported serovars in humans being Salmonella Enteritidis and S. enterica subsp. enterica serovar Typhimurium (S. Typhimurium). The majority of reported outbreaks of S. Typhimurium are considered to be associated with foodborne transmission [3]. Since 2000, vehicles of reported S. Typhimurium outbreaks have included: lettuce (n = 3), duck eggs (n = 2), cooked ham (n = 1), pre-packaged salad (n = 1), pre-packaged sandwiches (n = 1), lamb (n = 1), suspected raw lamb liver (n = 1), and chocolate products (n = 1) [2, Reference Harker4–Reference Larkin6].
Whole genome sequencing (WGS) of all Salmonella isolates received by the UK Health Security Agency (UKHSA) (formerly Public Health England) was implemented in April 2014, which improved the detection of outbreaks of gastrointestinal illness (GI) [Reference Ashton7, Reference Dallman8]. Isolates are grouped using a single-linkage clustering approach based upon single-nucleotide polymorphism (SNP) differences, enabling the identification of linked cases even when epidemiological links are obscured [Reference Ashton7–Reference Waldram9]. A 5-SNP threshold denotes high genetic relatedness and identifies cases likely associated with a common source [Reference Dallman8, Reference Waldram9].
Point-source outbreak, Cardiff, Wales
A previously described outbreak of 22 cases of genetically related S. Typhimurium was detected in Cardiff, Wales, in July 2021 following an Eid al-Adha barbecue celebration [Reference Adamson, Sawyer and Hobson5]. Isolates belonging to the same 5-SNP single-linkage cluster were first detected in Mid Wales in April 2018 and intermittently thereafter [Reference Adamson, Sawyer and Hobson5]. The clinical severity of cases in the barbecue outbreak was high, with 55% of cases reporting attendance at emergency health services and 27% requiring admission [Reference Adamson, Sawyer and Hobson5]. Contaminated sheep meat and lamb liver consumed raw were identified as likely vehicles of infection.
Wider investigations, UK
Further S. Typhimurium infections belonging to the same 5-SNP single-linkage cluster to those in the point-source barbecue outbreak were subsequently identified. Here, we describe investigations into the national expansion of the cluster of S. Typhimurium that was linked with ruminant livestock and related food chains in the UK.
Methods
Epidemiological investigation
Descriptive study
A case was defined as a resident in the UK with laboratory-confirmed S. Typhimurium belonging to the same 5-SNP single-linkage cluster as the barbecue outbreak strain according to the UKHSA SNP pipeline and with a specimen date between 1 August 2021 and 31 December 2022. Barbecue attendees were excluded from analyses given their known source of infection [Reference Adamson, Sawyer and Hobson5].
For cases in England and Wales, routine surveillance data collected by the UKHSA Gastrointestinal Bacteria Reference Unit were used to obtain demographic and laboratory information (date of data extraction: 3 January 2023). Environmental Health Practitioners and Health Protection teams collected questionnaire information via telephone interview for all cases in Wales (as is routine) and a subset of cases in England (not routine) on exposures in the week before symptom onset, or positive specimen date if the former was missing. Data were collected on symptoms, hospital admission, travel, contacts, food history, household water supplier, pets, contact with animals and/or birds, swimming, and contact with faeces. Trawling questionnaires were used by national teams in the UKHSA and Public Health Wales (PHW) for a subset of cases using purposive sampling, prioritizing by most recent date of symptom onset, to obtain further exposure information via telephone interview. Trawling questionnaires were used to generate hypotheses pertaining to potential vehicles of infection and premises for investigation.
For cases in Scotland, routine surveillance data collected by Public Health Scotland were used to obtain demographic and laboratory information (date of data extraction: 17 March 2023).
Intelligence on localized outbreaks in England was received through personal communication with incident directors in regional UKHSA teams.
Case–case study
PHW compared exposures in cases from Wales with those of individuals notified with non-Typhimurium Salmonella (defined as ‘case–controls’) [Reference Harker4] with sample date between 1 August 2021 and 16 May 2022 to test associations between case status and exposures. The ratio of case–controls to cases was 3:1. Case–controls were selected using simple random sampling. Secondary cases (defined as onset of illness ≥24 h after illness in a household with a primary case, or close contact with an individual reporting diarrhoea or vomiting) and cases who had travelled abroad in the week prior to symptom onset were excluded.
For cases, information from routine or trawling questionnaires was supplemented using an enhanced questionnaire which was based on the findings of trawling questionnaires. Interviews were conducted by the PHW Field Epidemiology Team by telephone and included detailed questions relating to the week prior to the earliest of symptom onset or specimen date. For case–controls, data were obtained from routine questionnaires on individuals with reported Salmonella infection (response rate: 50%).
Data analysis
Cases were summarized by time, place, and person. Continuous data were assessed for normality using histograms and the Shapiro–Wilk test [Reference Shapiro and Wilk10]. Skewed data were summarized with the median and interquartile range (IQR). Categorical variables were presented in frequency tables and missing data summarized. Pearson’s chi-squared [Reference Pearson11], Fisher’s exact [Reference Fisher12] and Wilcoxon rank-sum tests [Reference Mann and Whitney13] were used to compare differences in all cases and trawled cases, and cases and case–controls by demographic and clinical variables to assess comparability. Exposure attack rates were calculated.
Odds of exposures among cases and case–controls with 95% confidence intervals (95%CI) were calculated using logistic regression. Observations with missing values were excluded from statistical analysis. Stratified analysis was used to assess for potential confounding and effect modification. A multivariable model was created using forward-stepwise logistic regression, with each variable retained if significant (p < 0.05) in the likelihood ratio test [Reference Harrell14].
Analysis was carried out using Stata v14.2 [15], and visualizations were created using RStudio v4.2.2 [16].
Environmental and microbiological investigations
Tracing supply chains and environmental sampling
Environmental Health Practitioners investigated selected premises (e.g., butchers, restaurants, cafés) reported by ≥2 cases in questionnaires. The Food Standards Agency (FSA) then made enquiries to trace supply chains to source (e.g., abattoirs). Suppliers were prioritized for inspection by the FSA if supplying ≥2 premises under investigation. Environmental swabs were taken from implicated suppliers (e.g., equipment and carcasses). Specimens were cultured for bacterial pathogens including Salmonella spp., and all Salmonella isolates were typed and sent to the Gastrointestinal Bacteria Reference Unit for confirmation and sequencing. Information on routine microbiological sampling of carcasses (in accordance with Regulation (EC) No 2073/2005 [17]) undertaken by food business operators at abattoirs and corrective enforcement action was provided by the FSA.
Animals and animal environments
The Animal and Plant Health Agency (APHA) surveillance system comprises diagnostic, serotyping, and WGS data from animal and post-mortem samples submitted as part of disease investigations, described in detail elsewhere [18]. Any Salmonella identified in species for which Salmonella is reportable under the zoonosis order is received at the Salmonella National Reference Laboratory at APHA Weybridge for further characterization by WGS and, on occasion, by conventional serotyping methods in parallel.
Data were obtained from the APHA surveillance system for samples with specimen date between 1 August 2021 and 31 December 2022.
Whole genome sequencing (WGS) and phylogenetic analysis
Isolates were sequenced on an Illumina sequencing platform, as described previously [Reference Chattaway19, Reference Dallman20]. Sequences were analysed to determine the sequence type, serotype, and SNP type.
Illumina reads were mapped to S. Typhimurium reference genome (AE006468) using BWA v0.7.12 and Samtools v1.1 [Reference Li and Durbin21, Reference Li22]. High-quality variants (SNPs) were identified using GATK v2.6.5 in unified genotyper mode [Reference McKenna23]. High-quality core-genome SNPs (>90% consensus, minimum depth 10×, GQ ≥30) were extracted from SnapperDB v0.2.8 [Reference Dallman8]. IQtree v2.0.4 [Reference Minh24] was used to derive the maximum-likelihood phylogeny of the isolates. Visualization and annotation of the phylogeny was performed through the iTol platform.
Ethical approval
Ethical approval was not required as this study was conducted as part of an outbreak public health response. All data containing patient identifiable information was handled and stored in compliance with the Data Protection Act (2003) and GDPR (2018).
Results
Epidemiological investigation
Descriptive study
Routine surveillance
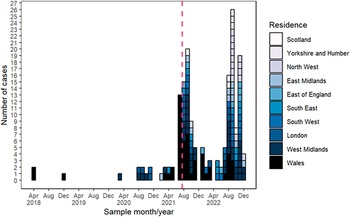
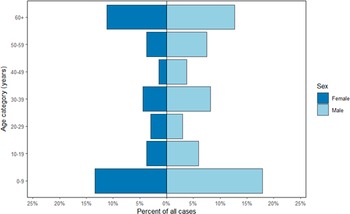
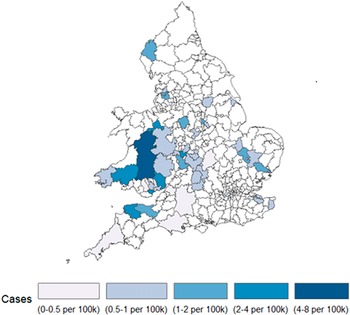
From 1 August 2021 to 31 December 2022, 142 cases were identified in the UK, of which 11 (8%) were known to be part of a localized outbreak associated with raw drinking milk in Northwest England between September to December 2022. Cases peaked in September following the point-source outbreak in July 2021 (Figure 1). The median age of cases was 32 (IQR: 5–57 years), and over half (59%, 81/137) were male (Figure 2). Cases were geographically dispersed across the UK, with 106 (/142, 75%) in England, 32 (23%) in Wales and four (3%) in Scotland. The incidence of cases was highest in Powys in Mid Wales (8 per 100,000 population) (Figure 3). Among cases with available information, fewer than five (/31) in England were hospitalized, and seven (/32, 22%) in Wales. No deaths due to infection with Salmonella were reported.

Figure 1. Epidemic curve of all cases in Salmonella Typhimurium five-single-nucleotide polymorphism single-linkage cluster, England, Wales, and Scotland, Apr 2018-Dec 2022 (N = 170). Date of barbeque outbreak indicated by vertical pink dotted line.

Figure 2. Age group (years) and sex of cases in Salmonella Typhimurium five-single nucleotide polymorphism single-linkage cluster, England, Wales, and Scotland, Aug 2021-Dec 2022 (N = 137). Excludes cases with unknown sex (n = 5).

Figure 3. Incidence map of cases in Salmonella Typhimurium five-single-nucleotide polymorphism single-linkage cluster per 100,000 population by residential local authority, England and Wales, Aug 2021-Dec 2022 (N = 101). Excludes cases with insufficient address data (n = 41).
Trawling questionnaires
Between August and September 2021, 19 trawling questionnaires were completed: 11 for cases resident in England and eight in Wales. Compared to all cases, cases who completed a trawling questionnaire were younger (median: 5 years, p = 0.04). The majority were of White (9/19, 47%) or (8/19, 42%) Asian British ethnicity. Median symptom duration was 14 days (IQR: 10–28 days).
Eating any meat and eating outside of the home were commonly reported exposures (both 15/19, 79%). Chicken (14/19, 74%) was consumed by most cases while lamb and mutton were reportedly eaten by few (<5/19, <26%). Approximately half of cases reported washing raw meats in their household (11/19, 58%) and adhering to a halal diet (i.e., meat processed as prescribed in accordance with the Islamic faith) (9/19, 47%).
Epidemiological investigation
Analytical study
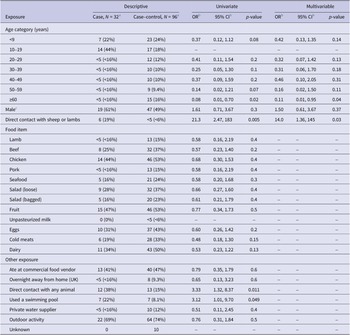
Among cases in Wales, 32 cases were eligible for inclusion in the analytical study. Cases (median: 14 years, IQR: 3–44) were younger on average than case–controls (median: 38 years, IQR: 16–58, p < 0.03) (Table 1). Most cases were male (19/31, 61%) but no statistical difference by sex was observed compared to case–controls (p = 0.3). Cases were more frequently identified in Cardiff (10/32, 31%), the capital city of Wales (19/86, 20%, p = 0.001), than case–controls. Case–controls had infection with Salmonella Enteritidis (30/96, 31%), Infantis (21/96, 22%) or one of 28 other serovars. Fewer case–controls reported bloody stools (6/86, 7% of case–controls vs. 8/28, 29% of cases, p < 0.01).
Table 1. Selected demographic, laboratory, and clinical characteristics of Salmonella non-Typhimurium controls (N = 96) and cases of Salmonella Typhimurium in five-single-nucleotide polymorphism single-linkage cluster (N = 32), Wales, Aug 2021–Dec 2022

a n (%).
p < 0.05
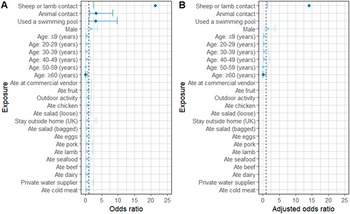
Exposure data were missing for up to 10% (10/96) of case–controls (Table 2). In univariate analysis, illness was associated with direct contact with sheep or lambs (OR: 21.3, 95%CI: 2.5–183), any direct animal contact (OR: 3.3, 95%CI: 1.3–8.4), swimming pool use (OR: 3.1, 95%CI: 1.01–9.7), and young age (≥60 vs. 10–19 years, OR: 0.08, 95%CI: 0.01–0.70). After adjusting for age and sex, the association between direct contact with sheep or lambs and illness remained (adjusted OR: 14.0, 95%CI: 1.4–145) but was reported by few (6/32 cases, 19%) (Figure 4).
Table 2. Univariate and multivariable analysis of selected exposures of cases in Salmonella Typhimurium five-single-nucleotide polymorphism single-linkage cluster (N = 32) and Salmonella non-Typhimurium case–controls (N = 96) reported in the week prior to symptom onset, Wales, Aug 2021–Dec 2022

a n (%).
b CI, confidence interval; OR, odds ratio.
c Excludes unknown gender (n = 1).

Figure 4. Forest plot of crude (a) and adjusted (b) odds of selected exposures reported by cases in Salmonella Typhimurium five-single-nucleotide polymorphism single-linkage cluster (N = 32) in the week prior to symptom onset compared to Salmonella non-Typhimurium case–controls (N = 96), Wales, Aug 2021-Dec 2022. Reference for age group is 10–19 years. Dark blue colour used to denote statistical significance (p < 0.05). Error bars not shown for 95% confidence intervals which include odds ratios >25.
Environmental and microbiological investigation
Tracing supply chains
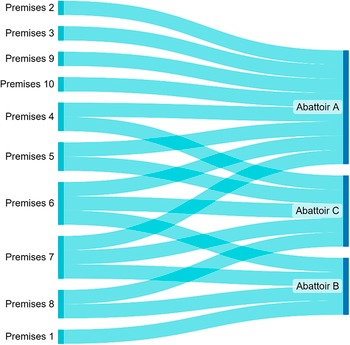
No single food item, premises, or supplier was identified as a common link between all cases. Livestock were identified to have usually passed from farms through markets, before being batched together with animals from other farms and taken to abattoirs for slaughter. Abattoirs frequently received livestock from different suppliers, some had changed ownership in recent years, and invoice trails were difficult to follow.
Three abattoirs (A, B, and C) were identified as suppliers to premises reported by ≥2 cases. Abattoir A was a large red meat slaughterhouse which supplied butchers throughout England and Wales. The FSA took investigation and enforcement action against this abattoir after poor hygiene practices were identified in July and August 2021, including unhygienic storage of tools, bunching of carcasses (which can result in cross-contamination), and high faecal contamination of carcasses. Of all investigated abattoirs, Abattoir A was the most common supplier to implicated premises, with known links to eight premises reported by cases (Figure 5). Abattoir B was another large red meat slaughterhouse operating in England. Four premises were linked to Abattoir B. Abattoir C was a small abattoir operating in Wales with links to five premises, which were all also supplied by Abattoir A (2), B (1), or A and B (2).

Figure 5. Flow diagram of food supply chain trace back investigations for premises reported by cases in S. Typhimurium five-single-nucleotide polymorphism single-linkage cluster, England and Wales, 2021.
Animals and animal environment sampling
S. Typhimurium isolates within the 5-SNP single-linkage cluster were identified in 47 samples from 36 premises in England and Wales submitted to the APHA for diagnostic and monitoring purposes. This included 20 samples from cattle, nine from sheep, eight from chickens, five from dogs and one from a swan. Three samples of raw pet food and one sample of mixed oil seeds intended for poultry feed were also identified in the same 5-SNP cluster.
Environmental sampling
Environmental samples from Abattoir A and B were positive for Salmonella spp. but did not belong to the same 5-SNP single-linkage cluster. No sampling was undertaken at Abattoir C due to its low throughput.
An isolate belonging to the same 5-SNP cluster was identified in a sheep carcass sample in Abattoir D in England in December 2021. This isolate was collected as part of routine sampling.
Phylogenetic analysis
The 5-SNP single-linkage cluster reflected some diversity (Figure 6). Highly genetically related clusters were observed for previously reported incidents, indicating common sources of infection. Isolates from animal, food, and human samples were distributed across the phylogeny.

Figure 6. Maximum-likelihood phylogenetic tree of S. Typhimurium isolates belonging to a common five-single nucleotide polymorphism single-linkage cluster (N = 265).
Discussion
We provide evidence of a highly genetically related cluster of S. Typhimurium among humans that was associated with ruminant livestock and related food chains in the UK. The evidence for this is threefold. Firstly, individuals who had contact with sheep or lambs were 14 times more likely to become infected compared to case–controls drawn from other Salmonella serovars. However, fewer than one in five cases reported this exposure and there were no obvious epidemiological links between the remaining, geographically dispersed cases. It is therefore likely that the cause of infection was not only ovine, as is consistent with the isolation of the outbreak strain from wide ranging samples, including wildlife, poultry, and animal environments. Secondly, the three abattoirs identified as suppliers to premises attended by multiple cases in trace back investigations were red meat slaughterhouses, one of which had enforcement action taken against them by the FSA for process hygiene failures between July–August 2021. Thirdly, an isolate in the same 5-SNP single-linkage cluster was identified from a sheep carcass sample at another abattoir in December 2021.
Control measures exist along the farm to fork pathway to limit zoonotic and foodborne transmission of pathogens from livestock via food products to people. Control failures which occur upstream in the food chain can cascade and increase the likelihood of contamination at multiple points later in the farm to fork pathway. In April 2018, the first cases in the S. Typhimurium cluster were detected in Mid Wales among individuals living on a sheep farm [Reference Adamson, Sawyer and Hobson5]. The Incident Management Team was informed through personal communication that then, in October 2020, an isolate belonging to the same 5-SNP single-linkage cluster was isolated from an employee at a lamb and mutton supplier, Abattoir D, in England. In December 2021, the strain was linked to this abattoir again after it was isolated from a sheep carcass. Previously, this abattoir had been implicated in a similar S. Typhimurium outbreak that was linked to cull ewes and investigated between July 2017 and August 2019 [Reference Carson and Davies25, 26]. While Abattoir D was not identified as a supplier to premises attended by cases in our trace back investigations, it is possible that connections were missed due to the complexity of the distribution network and unavailability of samples and detailed food histories for all cases. Similarly, isolates from samples taken from Abattoir A or B did not belong to the same 5-SNP single-linkage cluster. However, both abattoirs were linked to multiple premises identified in trace back investigations and Abattoir A had enforcement action taken against them during the study period for breaches in hygiene practices. These findings are indicative of upstream control measure failures and provide biologically plausible mechanisms for the potential amplification of contamination in the human food chain from ruminant products.
As well as connections to Abattoir D, our investigation had another important commonality with the 2017–2019 outbreak of S. Typhimurium [Reference Carson and Davies25]. As previously described [Reference Adamson, Sawyer and Hobson5], reported cases of S. Typhimurium increased in July 2021 after 22 individuals linked to an Eid al-Adha barbecue were exposed to contaminated sheep meat and a raw lamb liver dish. Eid al-Adha, meaning ‘festival of sacrifice’, is one of the most important festivals in the Muslim calendar [Reference Osman27]. In some countries, Muslims may sacrifice an animal for meat during Eid al-Adha, usually sheep [Reference Osman27]. In the S. Typhimurium outbreak in the UK that occurred between 2017–2019, rises in cases were linked to the cull ewe meat supply chain [Reference Carson and Davies25]. Historically, increased demand for cull ewes in the UK has been associated with the Muslim festival of Ramadan [28]. It is uncommon for S. Typhimurium to cause clinical disease in sheep, but host resistance can decrease and bacterial shedding can increase with stress and in the extremes of age [Reference Grunberg29]. Farms which mix and move large numbers of livestock, such as lambs or cull ewes, are said to be at high risk from Salmonella spp. and other diseases for this reason [30]. Supply chain and environmental contamination due to Salmonella spp. may therefore increase around the time of heightened demand for ruminant livestock which, when amplified by warmer summertime conditions, may increase the risk of an outbreak in humans.
Control measures
Animal production
The identification of isolates in the 5-SNP single-linkage cluster in numerous animal sources, including wildlife and poultry, was indicative of likely widespread environmental contamination and spread to other red meat sources. In response to the cluster, the APHA increased targeted communication to industry partners and the veterinary community to promote best practice in biosecurity. This included reinforcing the requirement that visibly unwell livestock should not be sent for slaughter. Producers were also reminded of their duty to correctly complete food chain information documentation [31].
Slaughter and processing
Regulation (EC) No 2073/2005 states that food business operators have a legal responsibility to ensure that unacceptable quantities of microorganisms are not present in foodstuffs intended for human consumption [17]. For products marketed as to be eaten cooked, such as mutton and lamb, low quantities of Salmonella spp. are deemed acceptable permitting the review of animal origin, operator process controls and slaughter hygiene. The FSA enforced this action in Abattoir A and D.
Preparation and consumption
Proportional action was required to reduce the bacterial load in the food chain, but action was also required in the processes thereafter to prevent further contamination, especially as not all cases in the cluster reported livestock contact or were knowingly exposed to raw meat products. Health promotion activities taken in response to the barbecue point-source outbreak are described in detail elsewhere [Reference Adamson, Sawyer and Hobson5]. Additional measures introduced at the level of households following the ongoing detection of cases included a social media campaign run by the FSA and PHW in 2022 promoting safe barbecuing. The campaign was informed by ongoing engagement between Environmental Health Practitioners and affected North African networks to identify appropriate modifications, such as language translation, to reach specific communities.
Onwards transmission
A ‘Warn and Inform’ communication was issued to Directors of Public Health, medical directors, and primary care clinicians in Wales to raise awareness of the cluster and re-iterate the need for stool samples for individuals presenting with diarrhoea. Laboratory staff in Wales conducted enhanced notification of cases indicative for S. Typhimurium to Health Protection Teams over the study period to rapidly alert them to possible cases. Additionally, a weekly report of trends in Salmonella spp. was developed by the PHW Field Epidemiology Team to monitor potential exceedances in the period surrounding Eid-al-Adha and other religious festivals, when demand for ruminant livestock products might have been heightened, in 2022. The intended effect was to improve early case detection and thus, reduce onwards person-to-person transmission.
Limitations
At the animal production level, we were not able to elucidate the extent of involved supply chains. Similarly, we did not identify a direct link throughout the farm to fork pathway. Challenges in identifying connections between the different levels of the supply chain were compounded by the limited availability of livestock samples for microbiological testing. No information was collected for the supply chain of foods consumed by case–controls and food supply chain information was also minimal for cases, both due to resource constraints and because specific address data for premises attended was often missing. Traceback investigations were therefore largely based on information provided in trawling questionnaires, for which 19 were completed. The detection of Salmonella spp. in sheep and carcass samples is often as part of clinical investigations of disease as, at time of writing, ruminants are not subject to national disease control plans in the same way that poultry are [18]. Abattoir sampling was likely particularly low during the study period as routine abattoir inspections had been paused during the COVID-19 pandemic, resulting in a higher workload as restrictions eased. There was also no sampling for Salmonella spp. at implicated premises because this was not deemed proportionate; multiple butchers and restaurants were reported so the source of infection was thought to be further upstream. Delay between symptom onset, sampling and sequencing results likely influenced cases’ ability to recall exposure histories, including foods consumed and premises visited, when questioned. To limit recall bias, cases were prioritized for questionnaires according to most recent symptom onset date. Nevertheless, these factors likely impaired our ability to trace the infection to potentially many sources.
Additionally, enhanced surveillance data were only available routinely for cases in Wales, reducing power for statistical analysis and representativeness. Case–controls were selected from individuals infected with a non-Typhimurium serovar of Salmonella and the data collection method for obtaining exposure information differed from that of cases. The use of cases as controls is considered an appropriate comparison group for some outbreaks of GI illness [Reference Harker4]. However, exposures usually associated with salmonellosis, particularly the more common serovars among case–controls (Salmonella Enteritidis and Salmonella Infantis), such as poultry and eggs, may have been over-represented in the case–control group. This could have biased associations towards the null. However, the aim of our study was to elucidate exposures that were different to other common exposures to Salmonella serovars, such as poultry, and it was assumed that using case–controls would limit recall bias given that they too had been ill.
Conclusion
WGS-based surveillance facilitated the identification of a likely association between UK ruminant livestock product production and a wider cluster of S. Typhimurium that would likely not have been linked through epidemiological investigation alone. The result was a highly complex and multifactorial investigation, through which sheep were identified as one of likely many sources of infection. Similarities with a previous incident were also identified during the investigation [Reference Carson and Davies25], including a potential association between periods of increased demand on the ruminant livestock food supply chain and GI illness. After implementing control measures along the farm to fork pathway and once the incidence of cases had stabilized at a low level, the incident was closed in May 2023.
A multi-agency workshop was convened in June 2023 to discuss commonalities in recent S. Typhimurium investigations linked to ruminant livestock food chains. Here, it was agreed that data sharing, such as of human and animal sequencing information, between agencies had greatly assisted outbreak investigations. It was therefore recommended that considerations be made to how these data can be shared proactively for routine surveillance to identify and respond to animal product-related outbreaks of human disease before they can escalate.
Data availability statement
The sequencing data that support the findings of this study are openly available in EnteroBase (https://enterobase.warwick.ac.uk), reference number [Reference Achtman32].
Acknowledgements
We wish to thank the Incident Management Team for their work on the point-source barbecue outbreak and wider investigations, including representatives from PHW, UKHSA, Public Health Scotland, FSA, APHA and Local Public Health Teams. We would also like to thank colleagues in Health Protection Teams and Environmental Health Practitioners who assisted with case investigation and interviews, John Cowden, and Robert Smith for their support in the Incident Management Team Epidemiology Subgroup, Liljana Petrovska and Derek Brown for whole-genome sequencing input and Mark McGivern for insights into localized outbreaks. Finally, we would like to thank individuals for taking the time to share their personal information for the purposes of this investigation.
Author contribution
R.A., S.M., and D.T. jointly chaired the Incident Management Team. D.T. led the Incident Management Team Epidemiology Subgroup. R.M., R.P., D.T., L.L., S.B., and C.W. designed the case–case study. C.P., J.M., L.L., and R.E. provided data for cases in England and Wales. L.B. provided data for cases in Scotland. A.M., J.L., and L.S. led and provided data on animal and animal environment investigations. C.Po., J.Li., T.P., and N.H. led and provided data on supply chain trace back and environmental investigations. K.T., D.L., and I.G. provided input on risk assessments for the Food Standards Agency. R.M. performed the data analysis. R.P. created the food supply chain flow diagram. A.P. created the phylogeny, with input from C.P., J.M., R.M., and L.S. R.M. and D.T. drafted the manuscript. A.P. drafted the whole-genome sequencing and phylogenetic analysis methods and results sections. All authors reviewed the manuscript and approved the final version.
Funding statement
This study received no specific grant from any funding agency, commercial, or not-for-profit sectors.
Competing interest
Authors R.P., P.S., L.F., C.S., N.P., R.S., C.W., R.A., S.M., and D.T. are employed by Public Health Wales. Authors R.M., C.P., L.L., S.B., A.P., J.M., and R.E. are employed by the UK Health Security Agency. Author L.B. is employed by Public Health Scotland. Authors A.M., J.L., and L.S. are employed by the Animal and Plant Health Agency. Authors C.Po., J.Li., T.P., N.H., K.T., D.L., and I.G. are employed by the Food Standards Agency.