INTRODUCTION
The protozoan parasites Cryptosporidium parvum and C. hominis are common causes of gastrointestinal disease worldwide [Reference Fayer, Fayer and Xiao1]. The infection can be transmitted by ingestion of water or food contaminated with oocysts, or by direct contact with infected faeces. While C. hominis is thought to cycle primarily among humans, C. parvum infects humans and young livestock and can be transmitted zoonotically. Cryptosporidiosis may manifest sporadically or as epidemic outbreaks. Numerous outbreaks resulting from person-to-person transmission, ingestion of contaminated water or food, or contact with farm animals (including in veterinary students) have been described (see e.g. [Reference Alpert2–Reference Valderrama20]). Traditionally, the most compelling reason to investigate these outbreaks has been to determine the source of infection, and most efforts have been directed towards the identification of exposure, rather than constitutional risk factors. Furthermore, many of these outbreaks involved open communities and were protracted, requiring the use of case-control studies for the investigation.
In 2006, we investigated an outbreak of gastrointestinal illness in a class of 96 veterinary students, in which C. parvum was identified as the sole enteropathogen in multiple faecal specimens. The molecular and epidemiological data converged in indicating a synchronous exposure of the class to C. parvum, which enabled the cohort study of exposure and constitutional risk factors reported in this paper.
MATERIALS AND METHODS
Sequence of events
On 10 October 2006 (day 14), a concerned student from a class of 96 enrolled in the Bachelor of Veterinary Science programme of Massey University, New Zealand, informed a co-author of this paper about the occurrence of several cases of illness, which could be linked to a practical class held on 26 September (day 0) in the Large Animal Teaching Unit (LATU), in which the class handled calves aged <1 month. The same reporting student stated that on 9 October (day 13), three students from the same class reported a gastrointestinal illness to a regional public health officer, and that faecal specimens from these students were submitted to a regional diagnostic laboratory. No other academic or recreational activities that could have resulted in exposure of the class to enteropathogens, or cases of gastroenteritis in students from other classes were recorded during the same period. During the practical class at LATU, the students stayed in a pen for about 20 min, to practise thoracic auscultation and rectal temperature and respiratory rate measurement in calves. They were wearing overalls and gumboots, and the gumboots were washed and overalls removed following the practical class. No gloves were provided, but students were able to wash hands with hot water and soap. According to one of the tutors, one of the calves had diarrhoea, but this could not be corroborated.
An investigation was required to meet New Zealand legislative requirements for Occupational Health and Safety and Public Health. On 11 October (day 15), a request to conduct the investigation was made, which was approved by the University's Human Ethics Committee on 13 October (day 17). On the same day of approval, the authors asked the class to submit stool specimens anonymously to the Institute of Veterinary, Animal and Biomedical Sciences (IVABS) of Massey University. Specimens were received during the weekend of 13–15 October and the diagnosis of cryptosporidosis was released to the class on 16 October (day 20). An environmental investigation at LATU and the farm from which the newborn calves were obtained was performed on 17 October (day 21). A first questionnaire (Q1) was issued to the class on 19 October (day 23). A second questionnaire (Q2) was issued on 25 May 2007.
Environmental investigation
The LATU received potable water supply from a 200-m deep non-artesian bore that was sanitized at the point of use using a 5 μm filter followed by an ultraviolet light system (Trojan UVMAX; Canada). A 100-l water sample was collected from one of the drinking water taps on the same day of the investigation and filtered on-site using a commercial filter (FiltaMax, Idexx Laboratories, USA), at a flow rate of 2 l/min. A 200-ml sample was also collected in a sterile bottle, placed on ice and transported to an environmental microbiology laboratory for analysis for the presence of Escherichia coli bacteria. The same day, the dairy farm that provided the calves for the practical class was visited. The farm was situated about 5 km from LATU. Faecal specimens from nine calves aged >30 days were submitted on ice to IVABS Unfortunately, the calves actually used in the class had been sold and could not be traced, and other calves aged <1 month were not present at the day of the visit as the calving season had ended.
Microbiological and molecular analysis
Seven faecal specimens submitted by students were analysed for the presence of enteropathogens. Faecal smears were examined microscopically for the presence of Cryptosporidium oocysts and Giardia cysts using a commercial immunofluorescence test kit (MeriFluor C/G, Meridian Bioscience, USA). The specimens were also tested for the presence of Salmonella spp., group A rotavirus, and norovirus genogroups 1 and 2. Culture for Salmonella was performed by direct inoculation of faecal samples onto xylose lysine deoxycholate (XLD) plates incubated at 37°C for 24 h, and by inoculation onto XLD plates after enrichment in selenite broth for 24 h at 37°C. Testing for group A rotavirus was performed using commercial strip tests according to the manufacturer's instructions (Rota-Strip, Coris BioConcept, Belgium). Testing for norovirus genogroups 1 and 2 was performed at the New Zealand norovirus reference laboratory (Institute of Environmental Science and Research, Porirua City) using routine real-time reverse-transcriptase PCR. In addition, testing for Campylobacter and Cryptosporidium was initially performed in three specimens using routine diagnostic procedures at the regional diagnostic laboratory associated with the public health service. The nine bovine specimens collected on the farm were tested for the presence of Cryptosporidium oocysts, as described above. The water sample was tested for the presence of Cryptosporidium oocysts as follows: the filter was taken apart and the foam disks were homogenized with elution buffer. The eluant was collected and centrifuged at 1500 g for 15 min. The pellet's volume was recorded and the supernatant aspirated to 5 ml. Water was added so the pellet volume was ⩽5%. A Cryptosporidium and Giardia immunomagnetic separation kit (Dynabeads® GC-Combo IMS kit, Invitrogen, USA) was used for the separation of the oocysts/cysts, in accordance with the manufacturer's specifications. The sample was then deposited onto a slide, stained with FITC-labelled anti-Cryptosporidium and anti-Giardia antibodies (AquaGlo™ G/C kit, Waterborne Inc., USA) and screened microscopically for the presence of oocysts/cysts using an epifluorescence microscope.
The Cryptosporidium-positive human faecal specimens and the nine bovine specimens were held in a refrigerator, with no preservatives. At the end of 2007, genomic DNA was extracted from the specimens: and the presence of Cryptosporidium parasites was assessed by sequence analysis of the 18S ribosomal RNA (18S rRNA) gene and the polymorphic regions of the sporozoite 60-kDa glycoprotein (GP60) and the 70 kDa heat-shock protein (HSP70) genes. The PCR and sequencing procedures were as previously described [Reference Grinberg21].
Questionnaires
The questionnaires were delivered by hand and anonymously completed by interested students at the end of a class. In order to facilitate the identification of the dates of illness, a calendar reporting the dates of the main academic activities held between 18 September and 18 October was incorporated in Q1. To minimize response bias, Q1 did not include questions directly addressing the calf-handling practical class. Thus, the rate of attendance and the source of drinking water (tap water, other source) consumed by students during the practical class could not be assessed by Q1, and were assessed in Q2. Due to the possibility of recall bias, Q2 was designed as a short questionnaire, which only included the following questions: (1) did you attend last year's practical class at LATU? (yes/no); (2) Did you drink tap water at LATU on that day? (yes/no); (3), Did you declare suffering from abdominal discomfort, diarrhoea, or vomiting in the previous questionnaire? (yes/no).
Analysis of data
The responses to Q1 and Q2 were used to elicit the demographics of the outbreak and describe the natural history of disease, estimate that attack rate, define the epidemic curve, and for a risk-factor analysis. The responses to Q2 were only used to estimate the rate of attendance at the practical class and the source of drinking water during the same practical. A case was defined as any student reporting having had abdominal discomfort and/or diarrhoea and/or vomiting between 18 September and 18 October 2006, in Q1. For the description of the demographics of the outbreak, the statistical significance of the differences between proportions was assessed using two-tailed Fisher's exact test. For the risk-factor analysis, all the students meeting the criteria for the definition of a case were considered cases, regardless of the date of onset of the illness. The strength of association between exposure or constitutional variables and the outcome binary variable (case/non-case) was assessed using the relative risk (RR), its probability under a null model of RR=1, and the 95% confidence interval. Independent variables used were nationality (New Zealander/non-New Zealander); background (rural/urban); history of previous physical interactions with ruminants (no interaction, <1 or 2 interactions per year, >1or 2 interactions per year); taking chronic medication (yes/no); attendance at the calf-handling class (yes/no); drinking tap water at LATU during the class (yes/no). The presence of interactions between gender and other independent variables was explored by stratification, followed by calculation of Wald's statistic for heterogeneity of strata [Reference Rothman, Greenland and Lash22]. Adjusted Mantel–Haenszel relative risks [Reference Mantel and Haenszel23] were used to aggregate the results for those strata not showing heterogeneity (as indicated by Wald's test P>0·05). Calculations were performed using Rothman's Episheet software (http://members.aol.com/krothman/episheet.xls, downloaded 27 August 2005). The modest sample size of the cohort precluded multivariate analyses.
RESULTS
Response of the class
Overall, seven students submitted stool specimens to IVABS between 14 and 15 October. Three formed specimens had been collected on 9 October and initially sent to the regional diagnostic laboratory. These specimens were reclaimed from the laboratory by the same students, and resubmitted to IVABS. One liquid faecal specimen had been collected by another student on 8 October and refrigerated by the same student until submission. Three additional students submitted formed specimens with no accompanying information. A total of 80/96 (83%) students completed Q1, and 64 (67%) completed Q2.
Microbiological results
The three specimens initially analysed at the regional diagnostic laboratory tested negative for Campylobacter and one tested positive for Cryptosporidium. All the seven specimens submitted to IVABS tested negative for rotavirus, Salmonella spp., norovirus and Giardia, and four tested positive for Cryptosporidium oocysts by immunofluorescence. One Cryptosporidium-positive specimen had previously tested positive and one tested negative at the regional diagnostic laboratory. One Cryptosporidium-positive specimen had been collected on 8 October, two on 9 October, and one on 14 October. Only 3/4 Cryptosporidium-positive human specimens yielded editable 18S rRNA sequences. Two isolates had sequences identical to the C. parvum ‘Type A’ 18S rRNA gene sequence (GenBank accession number AF093490), and the third isolate had a sequence identical to the ‘Type B’ copy of the same gene [Reference LeBlancq24]. Only two Cryptosporidium-positive human specimens yielded clear GP60 sequences, which were identical with each other and belonged to the allelic group IIa A21G4R1 according to the nomenclature proposed by Sulaiman et al. [Reference Sulaiman25]. The same sequence has been previously seen in only one human C. parvum isolate in New Zealand (data not shown), and most recently in two human C. parvum isolates in Australia (GenBank accession number GU214363). All the four Cryptosporidium-positive specimens yielded clear HSP70 gene sequences. There were two different sequences at this locus. One sequence was present in three isolates and was identical to the corresponding segment of the HSP70 sequence of C. parvum deposited in GenBank under accession number UG9698. The second HSP70 sequence was present in one isolate, and differed from the previous sequence by an insertion of a thymine between cytosine (position 2088) and adenine (position 2089). The PCR of this apparently polymorphic isolate was not repeated, but the same polymorphism was conserved after re-editing the sequence, arguing against an editing error.
All the bovine specimens tested negative for Cryptosporidium oocysts by immunofluorescence. However, the DNA extracted from one specimen yielded a sequence by PCR, which was 99% similar to the 18S rRNA gene sequence of Cryptosporidium bovis (see [Reference Santin26] and GenBank accession no. AY741305). An identical sequence was not retrieved in Genbank. No Cryptosporidium oocysts were revealed and no E. coli were isolated from drinking water collected at LATU.
Demographic characteristics and clinico-epidemiological features
The demographic characteristics of the class are reported in Table 1. Seventy percent of the respondents to Q1 were female, and 28% were non-New Zealand citizens, reflecting the demographics of the entire class roll (data not shown). The mean and range of the students' ages were very similar in both genders, and there was no significant difference between the proportions of males and females coming from rural or urban backgrounds (P=0·3), or males and females that had never handled ruminants prior to the practical class (P=0·19). However, a greater proportion of males than females had previously handled ruminants more than once or twice a year (P=0·009). In both genders, the proportion of students that had previously handled ruminants more than once or twice a year was greater in students from a rural background (data not shown). There were 25/80 cases among respondents to Q1 (31% attack rate), and 15/64 among respondents to Q2 (P=0·35). Nine out of 25 cases (36%) sought medical advice during the course of the illness. All 64 respondents to Q2 had attended the calf-handling practical (Table 2), and none of them responded as having drunk tap water at LATU during the same practical, arguing against a water-borne source of infection there (Table 2). Sixteen out of 25 cases (64%) reported diarrhoea and abdominal discomfort in Q1; six (24%) abdominal discomfort, diarrhoea, and vomiting; two (8%) abdominal discomfort only, and one (4%) vomiting only. All the cases reported a self-limiting illness, with a median duration of 5–6 days and range of 2–23 days (Fig. 1). The epidemic curve is reported in Figure 2. Only two cases reported an onset of illness before the date of the practical class, and the median and mode of the reported date of onset coincided at 5 days after the practical class.

Fig. 1. Duration of illness in the outbreak of cryptosporidiosis, as elicited from the responses to questionnaire Q1. Due to missing answers only 20/25 cases are reported.

Fig. 2. Epidemic curve of the outbreak of cryptosporidiosis as elicited by the responses to questionnaire Q1. Due to missing answers, only 23/25 cases are reported.
Table 1. Demographic characteristics of the outbreak of cryptosporidiosis as elicited from the responses to questionnaire Q1, stratified by gender (denominators differ due to missing answers)
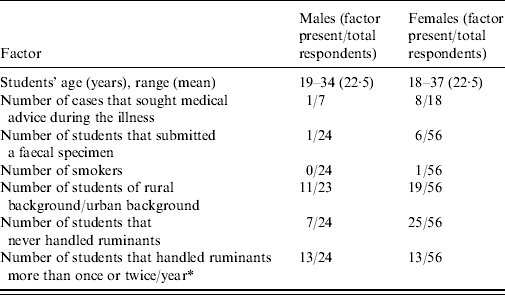
* Comparison resulting in a Fisher's exact test P<0·01.
Table 2. Risk-factor analysis of the outbreak of cryptosporidiosis, as elicited by the responses to questionnaires Q1 and Q2
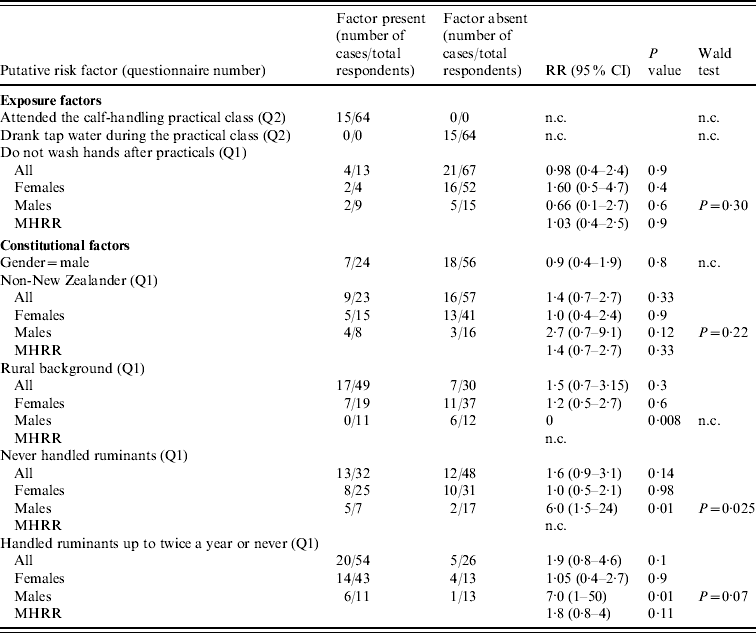
RR, Relative risk; MHRR, Mantel–Haenszel relative risk; CI, confidence intervals; n.c., not calculated due to a value of zero in the data, or because not applicable.
Denominators may vary due to missing data.
Risk-factor analysis
Table 2 shows the results of the risk-factor analysis. There were no differences in the proportion of cases between males and females (P=0·8), between New Zealanders and non-New Zealanders, neither overall (P=0·3), nor within each gender (P=0·9 in females, P=0·12 in males). In both genders, reporting not washing hands after practical classes was not associated with an increased risk of illness (P=0·4 in females, P=0·6 in males). No significant difference in the risk of illness was observed between students from rural or urban backgrounds (P=0·3). However, stratified analysis indicated a smaller risk in males from rural background compared to their urban counterparts (RR=0·000, P=0·008), and to rural and urban females (these relative risks can be extrapolated from Table 1). In agreement with this risk reduction in rural males, males that had never handled ruminants, or handled ruminants only once or twice a year prior to the practical class, were, respectively, six or seven times more likely to be cases, than males that handled ruminants more often (P=0·01). Such significant risk reduction was not observed in females (Table 2). Interestingly, 3/5 students who reported using chronic medication were cases and all three reported the use of anti-asthma inhaled steroids. Conversely, the two non-cases reported the use of drugs for other conditions (data not shown).
DISCUSSION
An outbreak of gastrointestinal illness occurred in a class of veterinary students. The identification of C. parvum as the sole agent from four faecal specimens provided evidence of an outbreak of cryptosporidiosis. The epidemiological evidence suggesting a synchronous exposure of the class to C. parvum enabled the collection of data on the incubation period and disease duration, and provided a rare opportunity to perform a risk-factor study using a prospective direction of enquiry. One of the main weaknesses of this study was the relatively small size of the cohort, which limits the generalizability of the results. Another limitation was the availability of only four laboratory-confirmed cases, which could have had implications for the specificity of the case definition. In fact, background cases unrelated to the outbreak may have been included in the analysis. However, the fact that only two cases with an onset before the day of the practical class were recorded (Fig. 2) suggested the presence of low level of background illness in the cohort.
The results of the subtyping supported the notion of a point-source outbreak, as simultaneous independent transmissions of C. parvum carrying the same rare IIa A21G4R1 allele would have been unlikely in New Zealand. The single nucleotide polymorphism observed in the HSP70 locus in one isolate was not due to an editing error, but may have reflected a PCR or sequencing error. These results, together with the circumstantial evidence and epidemic curve, converged in indicating an exposure to C. parvum during the practical class at LATU. Although a single negative water sample was of limited value to completely rule out a water-borne outbreak of cryptosporidiosis, Q2 indicated none of the respondents had drunk tap water during the practical class, leaving direct zoonotic transfer through contact with calves as the most likely route of transmission. Unfortunately, the source of infection could not be microbiologically traced to the farm of origin, perhaps because the period of shedding oocysts in calves is typically concentrated in the first 3 weeks of life [Reference Grinberg27], and such young animals were not present on the farm at the day of sampling.
An attack rate of 31% was estimated based on the responses to Q1. If the two cases reporting an onset of illness before the practical class are excluded, this rate is 29%. This might be considered high for an outbreak that is not water- or food-borne. However, experimental evidence indicates adult immunocompetent humans can be consistently infected with only 300 oocysts [Reference Chappell28–Reference DuPont31], and up to 107 oocysts/g have been reported in calves' faeces [Reference Grinberg27]. This means that only a speck of faecal material from an infected calf would be needed in order to infect a human.
If transmission during the practical class is assumed for this outbreak, the median incubation period is 5 days, with a range of 0–11 days (Fig. 2). These values are very similar to the results from experimental infections in volunteers, some of whom reported symptoms on the same day of the challenge [Reference Chappell28–Reference Okhuysen30]. The observed self-limiting diarrhoeal illness accompanied by abdominal discomfort and in a small proportion of cases, vomiting, is also similar to previous results [11, Reference Valderrama20, Reference Wolfson32, Reference Causer33]. One student reported as still suffering from the illness 23 days post-exposure. Later, the same student voluntarily reported suffering from repeated episodes of diarrhoea that eventually disappeared in January 2007 (data not shown). This case is consistent with a previous report by Hunter and colleagues [Reference Hunter34], of the occurrence of relapsing gastrointestinal symptoms in the course of cryptosporidiosis in immunocompetent hosts.
Overall, the type and duration of immunity conferred by naturally occurring C. parvum or C. hominis infections is not well understood. Newman et al. [Reference Newman35] found epidemiological evidence suggesting that symptomatic infections may result in the development of an immunity, and Chappell & Okhuysen hypothesized that immunity may develop after repeated exposures [Reference Chappell, Okhuysen, Smith and Thompson29]. However, the protection conferred by exposure to C. parvum has been studied in volunteers, with conflicting results. In one study, volunteers challenged with C. parvum showed partial resistance to the second challenge [Reference Okhuysen30]. Conversely, in another study, volunteers with pre-existing antibodies exhibited a more severe disease after an experimental challenge, than did their seronegative counterparts [Reference Chappell28]. In the present outbreak, a risk reduction in males coming from a rural background (compared to urban males and all females) was observed. This effect is consistent with the reduced risk also observed in males reporting having had previous contacts with ruminants (Table 2), as in New Zealand the rural environment is characterized by high livestock density. Immunity acquired through previous exposures to C. parvum as a result of contact with livestock, or other exposures in the rural environment in New Zealand, could plausibly explain the observed risk reduction. Unfortunately, our attempts to disentangle the effects of gender, rural or urban background, and frequency of handling ruminants on disease risk using multivariate analysis resulted in unbalanced designs due to the modest size of the cohort (data not shown).
In summary, the natural history of disease in this outbreak indicated a self-limiting illness of a median and mode incubation period of 5 days, corroborating data previously obtained from experimental infection trials in volunteers. A significant disease risk-reduction in males coming from a rural background, attributable to an immunity acquired through previous exposure to C. parvum in the farm environment, was observed. Interestingly, all students who reported chronic use of steroid inhalers for treatment of asthma were cases, indicating the need for studies of constitutional risk factors for cryptosporidiosis, other than immunosuppression [Reference Hunter and Nichols36]. This outbreak re-emphasizes the potential hazard for severe outbreaks of zoonotic cryptosporidiosis, and the need for hygiene when handling young calves.
ACKNOWLEDGEMENTS
We thank MidCentral District Health Board, New Zealand, for facilitating the testing for norovirus. This investigation was funded by Massey University and the New Zealand Ministry of Health.
DECLARATION OF INTEREST
None.






