1. Introduction
Episodes of mental illness may appear and disappear throughout a person's life, affecting around one in five individuals. Schizophrenia is a mental disorder affecting approximately 1% of the general population and is characterized by symptoms such as hallucinations, delusions, disorganized communication, poor planning, reduced motivation and blunted affect (Lewis & Lieberman, Reference Lewis and Lieberman2000; Lewis & Levitt, Reference Lewis and Levitt2002). Despite 50 years of research in schizophrenia, no effective approach for prevention or cure has been produced. The etiology of this disorder remains unclear, although research strongly points towards the interaction between genetic and environmental influences (Tsuang, Reference Tsuang2000; Aukes et al., Reference Aukes, Alizadeh, Sitskoorn, Selten, Sinke, Kemner, Ophoff and Kahn2008). Recent gene-expression and protein studies have reported an alteration of ubiquitin pathways in the brains from schizophrenia sufferers compared with controls (Bousman et al., Reference Bousman, Chana, Glatt, Chandler, Lucero, Tatro, May, Lohr, Kremen, Tsuang and Everall2010; Rubio et al., Reference Rubio, Wood, Haroutunian and Meador-Woodruff2013). The ubiquitin protein system plays an essential role in the regulation of membrane and cellular proteins, and has often been referred to as the “kiss of death” due to its labeling of proteins for degradation by proteases (Petroski, Reference Petroski2008; Tai & Schuman, Reference Tai and Schuman2008). The major function of the ubiquitin protein complex is to assure intracellular protein degradation by ubiquitination; a highly complex process involving a set of successive enzymes: E1 (ubiquitin activating enzymes), E2 (ubiquitin conjugating enzymes), E3 (ubiquitin protein ligases), the 20S proteasome and the deubiquitinating enzymes (Yi & Ehlers, Reference Yi and Ehlers2007). Ubiquitin is first activated by E1 ubiquitin activating enzymes, before being transferred to its active site, the amino acid cysteine. This transfer requires ATP, making the process energy dependent. The ubiquitin molecule is then passed on to the second group of enzymes within the complex, E2 (ubiquitin conjugating enzymes), before reaching the final group of enzymes, the E3 ubiquitin protein ligases, which recognize and bind the target substrate and label it with the ubiquitin. This process can be repeated until a short chain is formed, with three or more ubiquitin molecules usually targeting the protein to the proteasome, where the degradation occurs. Both E2 and E3 proteins exist as large families: more than 35 E2 s and 600 E3 s have been identified so far, resulting in highly complex combinations of E2 s with different E3 proteins defining the substrate specificity. Defects in this ubiquitin-dependent protein degradation have been implicated in the etiology of neurodegenerative diseases, metabolic disorders, cancer (Weathington & Mallampalli, Reference Weathington and Mallampalli2014), developmental deficiency, immunity pathologies (Sakamoto, Reference Sakamoto2002; Pagano & Benmaamar, Reference Pagano and Benmaamar2003) and schizophrenia.
We have focused our analysis on three genes coding for ubiquitin-related proteins (the ubiquitin conjugating enzyme E2D1 (UBE2D1), the E3 SUMO-protein ligase protein inhibitor of activated STAT 2 (PIAS2) and the E3 ubiquitin ligase F-box and leucine-rich repeat protein 21 (FBXL21), which is a SKP1-cullin-F-box protein, implicated in the regulation of the p53 pathway (Fig. 1). The p53 pathway plays an essential role in the modulation of neurodevelopmental processes (including cerebral vascularization and neurogenesis) and/or in neurodevelopmental disorders such as schizophrenia. Owing to their previous association in different reports and populations (Middleton et al., Reference Middleton, Mirnics, Pierri, Lewis and Levitt2002; Chen et al., Reference Chen, Wang, Sun, Chen, O'Neill, Walsh, Fanous and Kendler2008), we analyzed a set of potential single nucleotide polymorphisms (SNPs) in the UBE2D1 (coding for the E2D1 protein), PIAS2 (coding for the protein inhibitor of activated STAT 2) and FBXL21 (coding for the F-box and leucine-rich repeat protein 21) genes in the largest schizophrenia case–control Caucasian population collected in Australia in order to examine their potential associations with this devastating neurodevelopmental disorder.

Fig. 1. Schematic of the implication of the proteins coded by the genes of interest in brain development.
2. Materials and methods
(i) DNA samples
DNA samples were obtained from the Australian Schizophrenia Research Bank. Subjects with schizophrenia were identified using the Diagnostic and Statistical Manual of Mental Disorders IV criteria. All samples were from Caucasian volunteers. Subjects were matched for gender and age. The complete sample consisted of 268 schizophrenia cases, comprised of 186 males and 82 females, with an average age of 38·86 ± 11·01 years; and 268 matched controls, comprised of 169 males and 99 females, with an average age of 38·56 ± 12·57 years, with no prior history of mental disorders. After a complete description of the study to the subjects, written informed consent was obtained. This study was approved by and conducted according to the guidelines of the University of Wollongong Human Research Ethics Committee (HE10/161).
(ii) SNP genotyping
SNPs within the UBE2D1 (rs11006122 and rs1905455), PIAS2 (rs8094449, rs10502878, rs11876274 and rs56352844) and FBXL21 (rs1859427 and rs6861170) genes were tested in our Caucasian schizophrenia case–control population. The selection of these SNPs was based on their previous associations with schizophrenia and/or other disorders, and on their minor allele frequencies (MAF) reported in Caucasian populations (MAF > 15%) using HapMap data (http://hapmap.ncbi.nlm.nih.gov). High-throughput SNP genotyping was performed using the MassARRAY® genotyping assay (Sequenom, Inc., San Diego, CA, USA), with the analysis performed by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. PCR and extension primer design, selection and multiplexing were performed using MassARRAY® Designer Software (Sequenom, Inc., San Diego, CA, USA).
(iii) Statistical analysis
Power calculations indicated that for the smaller of the two cohorts, a sample size of 204 cases (408 alleles) with a MAF of 0·2 has >90% a priori power to detect a significant allelic association conferring an odds ratio of 1·5 or greater. The distribution of all tested SNPs did not deviate significantly from Hardy Weinberg equilibrium (p > 0·05), except for UBE2D1 (rs1905455, p = 1·88 × 10−17), which was then excluded from further analysis. To detect associations between each SNP and schizophrenia, χ 2 analysis was performed to test for significant differences in allele and genotype frequencies between the case and control groups. The significance for all statistical tests was set to p < 0·05 and p-values of <0·10 were described as trends. Data are expressed as specific counts for alleles and genotypes. Owing to multiple testing in the SNP analysis, a standard Bonferroni-corrected p-value of 0·007 (0·05/7) was required to give a 95% probability of correctly concluding not to reject the null hypothesis in the χ 2 test.
3. Results
Two SNPs within UBE2D1, four SNPs in PIAS2 and two SNPs within FBXL21 were analyzed for association with schizophrenia in a large Australian Caucasian population (268 schizophrenia patients vs. 268 matched controls with no prior history of mental disorders).
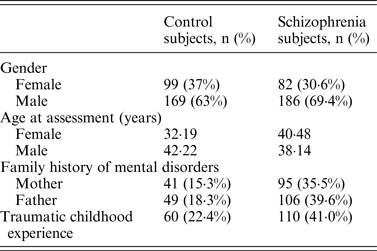
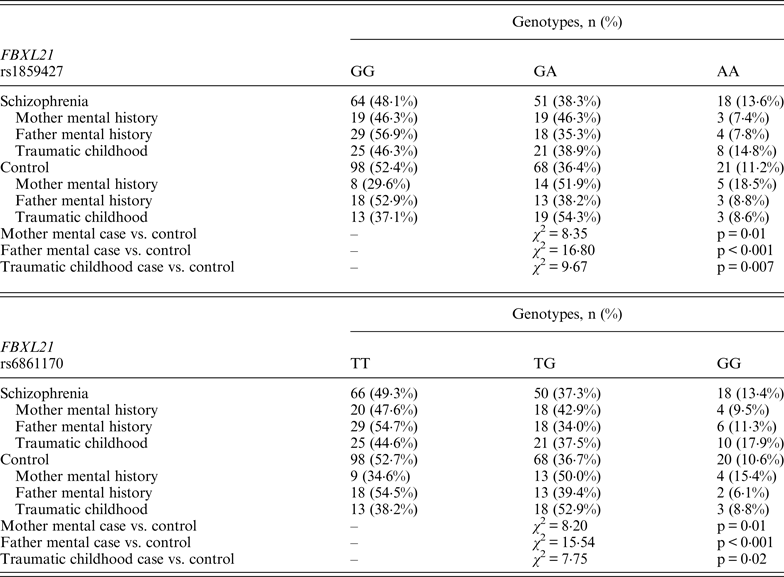
Following Bonferroni correction for multiple comparisons there were no significant associations between the allelic frequency of any of the tested SNPs (UBE2D1 rs11006122; PIAS2 rs8094449, rs10502878, rs11876274 and rs56352844; FBXL21 rs1859427 and rs6861170) and schizophrenia (0·08 ⩽ p ⩽ 0·92; Tables 1–3 respectively). In addition there were no genotypic associations between any of the tested SNPs and schizophrenia following Bonferroni correction (0·17 ⩽ p ⩽ 0·71; Tables 1–3). Furthermore, analysis of each of the genetic markers by gender revealed no significant allelic or genotypic associations with any of these genetic markers and either gender (0·08 ⩽ p ⩽ 1·00; Tables 1–3). Interestingly, the demographics of our tested population (Table 4) revealed that a large percentage of our schizophrenia subjects had a family history of mental disorders, twice as many as the control group. χ 2 analysis revealed that both of the FBXL21 genetic markers, rs1859427 and rs6861170, had a significant genotypic association with schizophrenia in the subjects whose mother (rs1859427: χ 2 = 8·35, p = 0·01; rs6861170: χ 2 = 8·20, p = 0·01) and/or father (rs1859427: χ 2 = 16·80, p < 0·001; rs6861170: χ 2 = 15·54, p < 0·001) had a history of mental disorders. None of the other SNPs from any of the other genes studied had any significant associations with schizophrenia in subjects who had a family history of mental disorders (0·10 ⩽ χ 2 ⩽ 4·17; 0·12 ⩽ p ⩽ 0·95). In addition, a large percentage of subjects from both tested groups experienced some form of self-reported childhood trauma, including but not limited to neglect, physical abuse, sexual abuse and post-traumatic stress, with a larger number of schizophrenia subjects (41·0%) having a traumatic childhood compared to the control group (22·4%). Again, χ 2 analysis revealed that both of the FBXL21 genetic markers, rs1859427 and rs6861170, had a significant genotypic association with schizophrenia in the subjects who experienced childhood trauma (rs1859427: χ 2 = 9·67, p = 0·007; rs6861170: χ 2 = 7·75, p = 0·02). Again, none of the other SNPs analyzed had any significant associations with schizophrenia in subjects who experienced trauma during childhood (1·49 ⩽ χ 2 ⩽ 4·34; 0·11 ⩽ p ⩽ 0·47).
Table 1. Allelic and genotypic distributions for UBE2D1 genetic marker in schizophrenia subjects and controls
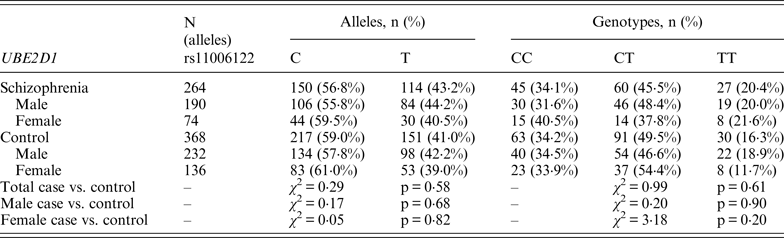
Table 2. Allelic and genotypic distributions for PIAS2 genetic markers in schizophrenia subjects and controls
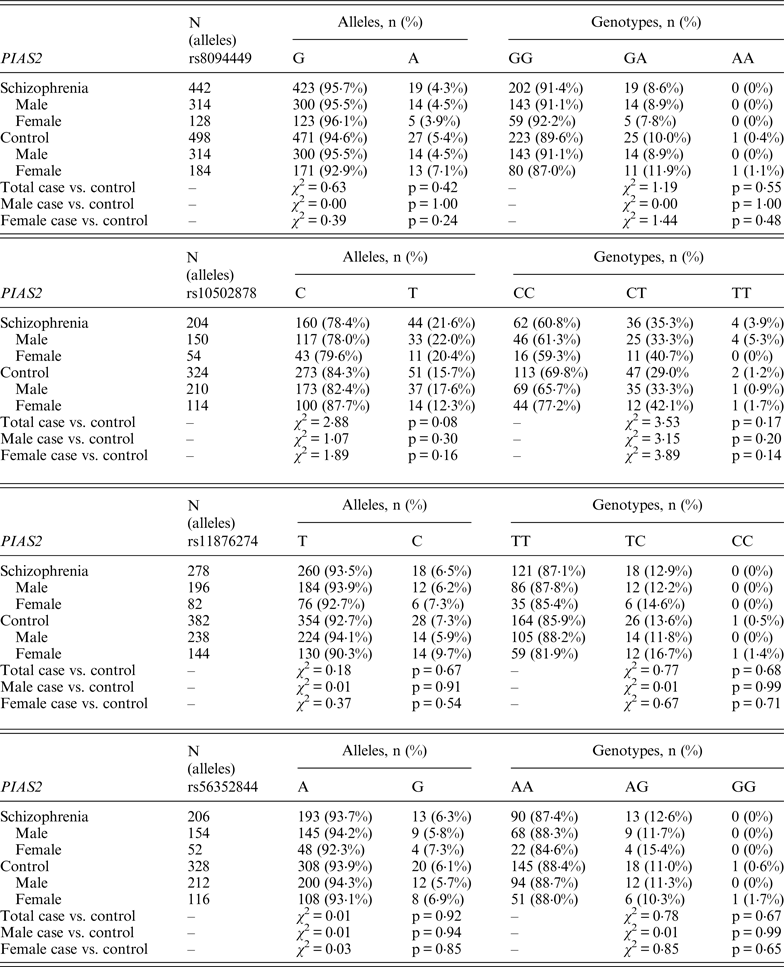
Table 3. Allelic and genotypic distributions for FBXL21 genetic markers in schizophrenia subjects and controls
Table 4. Demographics for control (n = 268) and schizophrenia subjects (n = 268)
4. Discussion
We have investigated the association of three genes from the ubiquitin protein system involved in the regulation of the p53 pathway in a large Australian Caucasian case–control schizophrenia population. We did not report any significant associations with schizophrenia for any of the tested SNPs in the UBE2D1, PIAS2 and FBXL21 genes in our population.
There is little information in the literature regarding association studies for our tested genes. Only the FBXL21 gene has been previously studied in two independent Irish populations, one corresponding to a high density of schizophrenia in Irish families (1350 subjects from 273 families) and the other was a large Irish case–control population (814 cases vs. 625 controls) (Chen et al., Reference Chen, Wang, Sun, Chen, O'Neill, Walsh, Fanous and Kendler2008). Chen et al., found rs1859427 and rs6861170 to be significantly associated with schizophrenia within their case–control population, and significance was maintained even after correction for multiple testing (p = 0·01967 for both markers) (Chen et al., Reference Chen, Wang, Sun, Chen, O'Neill, Walsh, Fanous and Kendler2008). The MAF for both rs1859427 and rs6861170 FBXL21 markers were very similar in the schizophrenia group for both the present study (MAF = 0·32 for both markers) and in the Chen et al. (Reference Chen, Wang, Sun, Chen, O'Neill, Walsh, Fanous and Kendler2008) study (MAF = 0·27). This suggests that there is likely to be a difference between the genotyping frequencies in the schizophrenia groups and/or frequencies in the control group between our study and the Chen et al. (Reference Chen, Wang, Sun, Chen, O'Neill, Walsh, Fanous and Kendler2008) study.
Interestingly, when we factor into our analysis the subjects who experienced some sort of trauma during childhood, the FBXL21 SNPs showed a significant association with schizophrenia among the tested SNPs (Table 5); however, none of the other tested SNPs in any of the other tested genes had any significant associations in schizophrenia subjects who self-reported any traumatic childhood experiences. Differential epigenetic gene regulation in relation to traumatic childhood experiences has only ever been reported for one ubiquitin E3 ligase (Mahogunin Ring Finger 1 or MGRN1) in women with fibromyalgia who had experienced traumatic childhood events (Menzies et al., Reference Menzies, Lyon, Archer, Zhou, Brumelle, Jones, Gao, York and Jackson-Cook2013). This suggests that adverse childhood experiences are able to induce genetic and epigenetic variations in ubiquitin protein genes responding to stressful conditions such as Ring Type E3 ubiquitin ligase and F-box proteins (Hermand, Reference Hermand2006; Hua & Vierstra, Reference Hua and Vierstra2011). A number of studies have shown strong associations between negative childhood experiences and adult psychiatric illnesses, in particular depression, psychosis and schizophrenia (Edwards et al., Reference Edwards, Holden, Felitti and Anda2003; Kelleher et al., Reference Kelleher, Harley, Lynch, Arseneault, Fitzpatrick and Cannon2008; Lu et al., Reference Lu, Mueser, Rosenberg and Jankowski2008). When we accounted for a family history of mental illness from either the mother and/or father within our tested population, we found a positive association between the FBXL21 SNPs and schizophrenia (Table 5); but again there were no significant associations with any of the other tested SNPs in the tested genes in schizophrenia subjects who had a family history of mental illness, suggesting an inheritance for the FBXL21 genetic markers in the context of psychiatric disorders. This is in line with the Chen et al. (Reference Chen, Wang, Sun, Chen, O'Neill, Walsh, Fanous and Kendler2008) study, which showed that the haplotype of FBXL21 markers rs1859472 and rs6861170 were over-transmitted in the Irish study of high-density schizophrenia families.
Table 5. Genotypic distributions for FBXL21 genetic markers in schizophrenia subjects and controls with respect to parental history of mental health issues and traumatic childhood experiences
As mentioned above, the FBXL21 gene encodes for an E3 ubiquitin ligase F-box protein in the SKP1-cullin-F-box complex. The FBXL21 protein is known to be able to stabilize cryptochrome (CRY) proteins CRY1 and CRY2, which are implicated in regulating mammalian circadian rhythms (Hirano et al., Reference Hirano, Yumimoto, Tsunematsu, Matsumoto, Oyama, Kozuka-Hata, Nakagawa, Lanjakornsiripan, Nakayama and Fukada2013). In fact, the FBXL21 protein was reported to antagonize the FBXL3 protein, another F-box-type E3 ligase, which ubiquitinates CRY proteins and mediates their degradation (Hirano et al., Reference Hirano, Yumimoto, Tsunematsu, Matsumoto, Oyama, Kozuka-Hata, Nakagawa, Lanjakornsiripan, Nakayama and Fukada2013). By attenuating the destabilizing action of FBXL3 on CRY proteins, FBXL21 expression allows for an adapted regulation of circadian rhythm gene transcription by CRY proteins according to the circadian cycle. Furthermore, the circadian pattern of FBXL21 expression in the mouse suprachiasmatic nucleus (region responsible for controlling circadian rhythm) is reminiscent of the expression pattern seen for other circadian pacemaker genes such as Period 1 (PER1) (Dardente et al., Reference Dardente, Mendoza, Fustin, Challet and Hazlerigg2008). Mutations in either the FBXL21 or FBXL3 genes can lead to a dysfunction of circadian rhythm oscillations and lead to significant behavioural disturbances in individuals and alterations in their sleeping patterns; moreover, the absence of FBXL21 causes a short-period phenotype in both mice and cells (Hirano et al., Reference Hirano, Yumimoto, Tsunematsu, Matsumoto, Oyama, Kozuka-Hata, Nakagawa, Lanjakornsiripan, Nakayama and Fukada2013; Yoo et al., Reference Yoo, Mohawk, Siepka, Shan, Huh, Hong, Kornblum, Kumar, Koike, Xu, Nussbaum, Liu, Chen, Chen, Green and Takahashi2013). Interestingly, schizophrenia patients have been reported to have intrinsically unstable circadian oscillators. A study by Bromundt et al., recently provided important new information concerning the link between impairments in neuropsychological function and disrupted circadian rhythms in schizophrenia (Bromundt et al., Reference Bromundt, Köster, Georgiev-Kill, Opwis, Wirz-Justice, Stoppe and Cajochen2011), which is further supported by a microarray study that found a significant downregulation of the circadian pacemaker gene PER1 in the postmortem temporal cortex of schizophrenia patients compared to healthy controls (Aston et al., Reference Aston, Jiang and Sokolov2004). Furthermore, an animal model study has shown dysfunction in the synchronization of circadian rhythms between brain cell networks involved in sleep–wake regulation and cognition (Dudley et al., Reference Dudley, Erbel-Sieler, Estill, Reick, Franken, Pitts and McKnight2003). Overall this suggests that alterations in circadian rhythms are present in schizophrenia and that polymorphisms in the FBXL21 gene, in addition to the FBX3 gene, could be involved in the circadian cycle disturbances that have been observed in schizophrenia (Mansour et al., Reference Mansour, Talkowski, Wood, Chowdari, McClain, Prasad, Montrose, Fagiolini, Friedman, Allen, Bowden, Calabrese, El-Mallakh, Escamilla, Faraone, Fossey, Gyulai, Loftis, Hauser, Ketter, Marangell, Miklowitz, Nierenberg, Patel, Sachs, Sklar, Smoller, Laird, Keshavan, Thase, Axelson, Birmaher, Lewis, Monk, Frank, Kupfer, Devlin and Nimgaonkar2009).
The FBXL21 gene is located on chromosome 5q31, a region that has previously been shown to contribute to the susceptibility for schizophrenia in both German and Israeli pedigree families (lod score 1·8) (Schwab et al., Reference Schwab, Eckstein, Hallmayer, Lerer, Albus, Borrmann, Lichtermann, Ertl, Maier and Wildenauer1997). Putative loci associated with psychosis in bipolar disorder pedigrees were characterized in the chromosomic region 18q21, which includes the locus for the PIAS2 gene (Park et al., Reference Park, Holzman and Goldman-Rakic1995). This region has also been suggested to be an influential genetic loci, common to both schizophrenia and bipolar disorders, depending on polygenic influence and critical environmental factors (Mors et al., Reference Mors, Ewald, Blackwood and Muir1997). Unfortunately, we did not report any significant associations between the tested SNPs (promoter and intron variants) in the PIAS2 gene with schizophrenia, suggesting that other genetic markers within this gene may be involved. Similarly, the genetic polymorphisms analyzed within the UBE2D1 gene (located in the promoter region) were not associated with schizophrenia in our tested population. The UBE2D1 gene is located at the chromosomic region 10q21·1 (http://www.ncbi.nlm.nih.gov/gene/7321). This region includes the Ankyrin 3 (ANK3) gene, previously associated with schizophrenia by a number of studies that found the rs10761482 SNP to be associated with schizophrenia in a large European population as well as in Han Chinese populations (Gella et al., Reference Gella, Segura, Durany, Pfuhlmann, Stöber and Gawlik2011; Yuan et al., Reference Yuan, Yi, Wang, Sun, Li, Du, Zhang, Yu, Fan, Li and Yu2012).
Owing to its early expression in brain development and its key role in genomic stability as well as in the apoptotic process in brain cells, the p53 pathway plays an essential role in the modulation of neurodevelopmental processes, including those within schizophrenia pathophysiology. Interestingly a reduced risk of cancer has been observed in individuals with schizophrenia during the last decade; considering the significant role p53 plays in the progression of cancer, this suggests a significant role of p53 in schizophrenia. Previous p53 polymorphisms were found to be associated with schizophrenia in both Chinese and Caucasian case–control populations and in family studies (Yang et al., Reference Yang, Xiao, Chen, Sang, Guan, Peng, Zhang, Gu, Qian, He, Qin, Li, Gu and He2004; Ni et al., Reference Ni, Trakalo, Valente, Azevedo, Pato, Pato and Kennedy2005). It is thought that the regulation of p53 expression by ubiquitin degradation may play an important role in schizophrenia pathophysiology. Although the present study did not examine polymorphisms within p53, all of the genes examined are involved in p53 signalling. While we did not find any significant associations between any of the tested polymorphisms and schizophrenia, replicating our study in a larger population and/or testing additional polymorphisms within the same genes will add to, and allow for further exploration into the role of the tested ubiquitin-related genes in the genetic vulnerability of schizophrenia.
Our study reports the analysis of potential SNPs in three candidate genes from the ubiquitin protein system in a large Australian case–control schizophrenia population. Owing to the limited information available on the role of E2 ubiquitin conjugating enzymes and E3 ubiquitin ligases in schizophrenia, in addition to the increasing variety of these groups of proteins, additional studies will be necessary to further examine the role of these ubiquitin proteins in the genetics of schizophrenia. A growing interest has recently emerged that is targeting ubiquitin-related proteins in the treatment of cancer and inflammatory-related diseases. Keeping in mind the paradoxical relationship between cancer and schizophrenia, the ubiquitin protein systems seem to be a good candidate for further study into the genetics and therapy of schizophrenia.
This work was supported by the Schizophrenia Research Institute, utilizing infrastructure and funding from NSW Health. This study was supported by the Australian Schizophrenia Research Bank, which is supported by the National Health and Medical Research Council of Australia, the Pratt Foundation, Ramsay Health Care, the Viertel Charitable Foundation and the Schizophrenia Research Institute. We thank Pavel Bitter and Shalima Nair from the Australian Cancer Research Foundation at The Garvan Institute of Medical Research in Sydney for their help in the MassARRAY genotyping assay of our samples. Jessica L. Andrews is supported by an Ian Scott Scholarship from Australian Rotary Health. This research received no specific grant from any funding agency, commercial or not-for-profit sectors.
Declaration of interest
None.








