INTRODUCTION
Group A rotaviruses are a leading cause of severe acute gastroenteritis (AGE) in children, and worldwide rotavirus-associated mortality was estimated to cause 37% of diarrhoea-related deaths in children aged <5 years in 2008 [Reference Tate1]. Since 2009, the World Health Organization has recommended that infants worldwide be vaccinated against rotaviruses [2], and the burden of the disease has substantially decreased in countries that have introduced vaccination [Reference Patel3].
The rotavirus genome is formed by 11 segments of double-stranded RNA that codify six viral structural proteins (VP) and six non-structural proteins. The external proteins VP7 and VP4, which are the main cause of the formation of neutralizing antibodies, are codified by two genetic segments that form the basis of the most widely used classification system (G-type [VP7] and P-type [VP4]) [Reference Santos and Hoshino4]. Currently, 27 G-types and 35 P-types of group A rotaviruses [Reference Matthijnssens5] are known. Reassortment is an important mechanism of rotavirus evolution, generating strains with different gene combinations [Reference Iturriza-Gómara, Isherwood, Desselberger and Gray6, Reference Maunula and Von Bonsdorff7]. Rotavirus G9, uncommon before 1995, was globally transmitted from the mid-1990s onwards, having been added to the major rotavirus genotypes G1–G4 [Reference Santos and Hoshino4, Reference Iturriza-Gómara8, Reference Linhares9]. G1–G4 and G9 cause more than 90% of rotavirus infections in Europe [Reference Santos and Hoshino4, Reference Iturriza-Gómara8].
The rotavirus vaccines Rotarix (GlaxoSmithKline Biologicals, Belgium), a live attenuated monovalent vaccine (G1[P8]) and RotaTeq (Merck & Co. Inc., USA), a pentavalent vaccine including strains from five human-bovine reassortant rotaviruses (G1–G4 and [P8]), became commercially available in Spain in 2006 and in 2007, respectively, but have not been included in the Basque Health System vaccination calendar, and are only dispensed in private practice. Based on the number of doses sold in Gipuzkoa, the mean coverage for both vaccines in the 2-year period 2009–2010 was estimated to be 11%.
Rotavirus G12 is considered an unusual genotype. However, due to its high capacity for reassortment, moderately increasing incidence and certain similarities with the evolution of G9 rotaviruses, there is concern that, in the future, this rotavirus could become a major human rotavirus genotype [Reference Rahman10–Reference Matthijnssens12]. The aim of this study was to describe a seasonal epidemic recently occurring in Gipuzkoa (northern Spain), in which the predominant genotype was rotavirus G12[P8].
MATERIAL AND METHODS
Study population
The epidemic was observed in the context of a prospective, population-based study performed from July 2009 to June 2011 in the counties of San Sebastián and Zumarraga (Gipuzkoa, autonomous region of the Basque Country), with 405 745 and 89 839 inhabitants, respectively (2006 census, Basque Institute of Statistics). The Donostia (San Sebastián, 77 paediatric beds) and Zumarraga (10 paediatric beds) hospitals cater for more than 97% of paediatric hospitalizations in the study area and are separated by 54 km. The study included all patients aged <3 years who sought medical care for AGE and for whom stool cultures were requested, both from the hospitals and from outpatient clinics. Duplicate episodes occurring in the same semester were excluded. Of the hospitalized children, infections in which the stool samples were obtained ⩾5 days after hospital admission were considered to be nosocomial.
Virological studies
The presence of group A rotavirus antigen in stool was investigated by an enzyme immunoassay (ELISA, ProsPecT™ Rotavirus kit, Oxoid Ltd, UK) or a chromatographic immunoassay (VIKIA® Rota-Adeno, bioMérieux SA, France). Positive samples were suspended in B199 medium (Sigma-Aldrich, USA), frozen at −80 °C and subsequently analysed for G- and P-type through multiplex, reverse transcription (RT)–polymerase chain reaction (PCR) methods [European Rotavirus Detection and Typing Methods version 4 (http://www.eurorota.net/docs.php)]. Viral RNA was obtained using the NucliSENS® Easy-Mag platform (bioMérieux). When no G- or P-type was detected, the presence of RNA of the VP6 gene was investigated by PCR [Reference Iturriza Gómara13]. Samples negative for this gene despite positivity in the antigen tests, were considered to lack sufficient viral RNA for genotyping, although we cannot exclude the possibility of some false-positive results of the antigen tests used. A sample of the outbreak-dominant genotypes and all unusual genotypes were sequenced using an AbiPrism 3100 Genetic Analyzer (Applied Biosystems, USA).
Statistical analysis
The χ2 test was used to compare proportions and analysis of variance (ANOVA) to compare the ages and length of hospital stay in the distinct groups of rotavirus-positive children. Significance was set at α = 0·05 (5%). Population-based incidence rates and hospitalization rates were calculated as described previously [Reference Cilla14].
RESULTS
In the study period, the presence of rotavirus was investigated in 4597 episodes of AGE and was found in 507 episodes, occurring in 504 children (294 boys and 210 girls). In both seasons (July 2009–June 2010 and July 2010–June 2011) there was a winter epidemic, reaching the maximum incidence in February (Fig. 1). Of the 507 episodes of rotavirus AGE, 85 (16·8%) were hospitalized, of which three required admission to the paediatric intensive care unit. There were no deaths. In 68 episodes (33 and 35 in each season), the main cause of hospital admission was acute dehydrating diarrhoea, and most admitted children (n = 61, 89·7%) required intravenous fluid replacement. In the remaining patients, rotavirus infection was considered incidental, either because the infection was acquired after admission (nosocomial infection, n = 9) or because it caused mild AGE and there was another cause of hospitalization (n = 6). In patients aged <2 years (454 episodes), admission rates were 30·6 [95% confidence interval (CI) 21·6–43·4] cases/10 000 children in the 2009–2010 season and 31·7 (95% CI 22·4–44·5) cases/10 000 children in the 2010–2011 season.

Fig. 1. Monthly distribution and dominant genotype of episodes of acute rotavirus gastroenteritis in children aged <3 years between July 2006 and June 2011 in Gipuzkoa, Basque Country, Spain (counties of San Sebastián and Zumarraga). The data for the period July 2006 to June 2009 are from reference [Reference Iturriza Gómara13].
Genotypes
A G- and/or P-type were obtained in 467 rotavirus strains, representing 92·1% of the total number of rotavirus AGE episodes (467/507) (Table 1). In the 2009–2010 season, genotype G9 was dominant, accounting for 81·6% (199/244) of genotyped strains. In the second season, the dominant genotype was G12, representing 65% of genotyped cases (145/223). G-type 12 was associated with P-type 8 [P8] in most cases (127/129).
Table 1. Rotavirus genotypes detected during two seasons in children aged <3 years from two counties in Gipuzkoa, Basque Country, northern Spain*
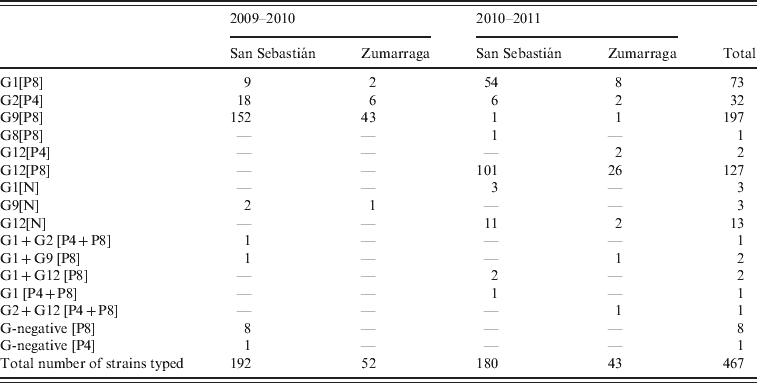
* In 20 samples, there was no remaining material for genotyping. In other 20 samples, no G or P-types were detected, and all these samples were also negative for VP6 RNA.
Characteristics of genotype G12 circulation in the 2010–2011 epidemic
Rotavirus G12 was detected in the 2010–2011 epidemic for seven consecutive months (September–March) in the area under surveillance, without any G12 strain having been detected in the preceding epidemic. The incidence of virologically confirmed rotavirus G12 infection in children aged <3 years was 93·6 (95% CI 78·0–112·3) and was 98·0 (95% CI 68·5–140·4) cases/10 000 inhabitants in the counties of San Sebastián and Zumarraga, respectively (χ2 = 0·05, P = n.s.). No significant differences were detected when the population was divided into persons living in rural settings with <1000 inhabitants, rural settings with 1000–10 000 inhabitants or urban areas with >10 000 inhabitants.
By age group, rotavirus G12 infection was found in 14 infants aged <4 months (9·7%), 61 infants aged 4 to <12 months (42·1%), 59 children aged 12 to <24 months (40·7%) and 11 children aged 24 to <36 months (7·6%). The mean age of patients infected with rotavirus G12 was 12·7 ± 7·0 months, similar to that of those infected with rotavirus G9 (13·2 ± 6·6 months) or G1 (13·8 ± 7·4 months) (ANOVA: F = 0·99, P = n.s.). Excluding mixed rotavirus infections, a total of 16·2% (23/142) of patients with rotavirus G12 were admitted to hospital due to community-acquired AGE, this percentage being 15·8% (12/76) and 14·5% (29/200) in patients with G1 and G9 infection, respectively (χ2 test with 2 d.f. = 0·25, P = n.s.). Of the G12 cases, we found no significant differences in age, sex, origin (rural or urban) or month of occurrence between hospitalized and non-hospitalized children.
The mean length of hospital stay in patients admitted for AGE caused by the three circulating rotavirus genotypes was similar (G1: 4·4 ± 2·5; G9: 4·3 ± 1·6; G12: 4·1 ± 2·8 days) (ANOVA: F = 0·09, P = n.s.). G12 caused nine episodes of nosocomial infection. Of the 35 children with rotavirus AGE with known rotavirus vaccination status in the 2010–2011 season (27 infected with G12, of which six were hospitalized), none had been vaccinated.
Molecular analysis of G12 strains
All G-type 12 strains that were sequenced (n = 47), including the two G12[P4] strains, belonged to lineage III (Fig. 2). The VP7 nucleotide sequences of the G12 strains found in 2010–2011 in Gipuzkoa showed very high similarity (99·7–100%), while the similarity with other G12 strains detected in 2004–2005 (96·7%) was somewhat lower. A BLAST search found that the strains were very similar (>99%) to G12 strains detected previously in Thailand, Sri Lanka, eastern India or the USA, but were only 90% similar to the prototype G12 strain L26. The VP4 gene sequences of nine G12[P8] strains found in this study showed 99·8% identity with each other, and exhibited a close relationship to that of a G9[P8] strain circulating in the USA in 2009 (Bethesda DC3, accession no. HQ702221) (99·2%), and to a G9[P8] strain detected in Gipuzkoa in 2011 (accession no. JQ410051) (98%). Partial VP7 and VP4 nucleotide sequences of the G12 rotaviruses found in this study were deposited in the GenBank database under accession nos. JQ410021–JQ410051.

Fig. 2. Phylogenetic analysis of 47 nucleotide sequences of G12 rotaviruses (region VP7, positions 48-870) from Gipuzkoa, Basque Country, Spain, July 2010–June 2011. A closed circle (•) indicates the rotavirus strains in this study and a triangle (▴) indicates G12 rotavirus strains detected in previous seasons (region VP7, positions 505-840). The sequences used for comparison were obtained from the GenBank database. The tree was constructed through the neighbour-joining method with 1000 bootstrap replications and shows bootstrap values higher than 70 in the branches. The distance is expressed as the number of the nucleotide substitutions per site.
DISCUSSION
The present report documents the characteristics of two seasonal rotavirus epidemics in Gipuzkoa and shows that in the 2010–2011 epidemic, a strain with an unusual G-P combination, G12[P8], predominated, being found in 65% of genotyped strains. Since genotype surveillance began in 1996, only four cases of rotavirus G12 infection had previously been detected, all between December 2004 and December 2005 [Reference Cilla15]. The characteristics of the 2010–2011 rotavirus AGE epidemic reproduced those of previous epidemics, with a similar seasonal pattern and affected age group, prolonged duration (7 months) and broad geographical spread. The impact of this epidemic was high, the rate of hospitalization being similar to that observed in previous seasons, in which rotavirus G1 or G9 were dominant [Reference Cilla14].
G12 rotaviruses were first detected in children aged <2 years with diarrhoea in 1987–1988 in the Philippines [Reference Taniguchi16], and no new strains were reported until 1998–1999 in Thailand [Reference Pongsuwanna17], the USA [Reference Griffin18] and Argentina [Reference Castello19]. In the first few years of this century, strains of this genotype were detected with a marked frequency in countries of the Indian subcontinent, such as Nepal (Kathmandu, 33% in 2003–2007), India (Delhi, 14·4% in 2000–2007), and Bangladesh (Dhaka, 9·6% in 2005–2006) [Reference Sherchand20–Reference Ahmed22], this Asian region possibly being the origin of G12 rotaviruses [Reference Rahman10]. In Europe, the first G12 rotaviruses were detected sporadically in the UK (2002) and Belgium (2003), and showed strong similarities with strains from Bangladesh [Reference Rahman10]. Between 2004 and 2006, strains were also detected sporadically in other European countries, including Spain, France and Slovenia [Reference Cilla15, Reference de Rougemont23, Reference Steyer24]. In Hungary, G12[P8] strains represented 6·9% of those genotyped in 2005 [Reference Bányai25]. G12 strains were found in only 0·2% of those genotyped in a survey of viral diarrhoea conducted in 2006–08 in Spain [Reference Sánchez-Fauquier26]. In a study performed in western Europe in 2004–2005, G12 strains were only detected in two of seven participating countries (Italy and Sweden) [Reference Van Damme27] and, although G12 strains were found in 15 of 16 participating countries in the European Rotavirus Network (EuroRotaNet) in 2006–2009, the incidence only exceeded 5% in one country (Finland, incidence 6·8%) [Reference Iturriza-Gómara8].
Due to the increasing tendency in the incidence of G12 rotaviruses, this genotype is currently classified as an emerging genotype [Reference Iturriza-Gómara8, Reference de Rougemont23] and may become a predominant genotype in the future [Reference Rahman10]. To our knowledge, outside the Indian subcontinent [Reference Sherchand20, Reference Sharma21], a seasonal epidemic in which rotavirus G12 predominated has only been reported in Rochester (New York, USA) in 2007 [Reference Payne28], and in a study conducted in 2008–2009 in Argentina [Reference Stupka29]. During this survey, G12[P8] strains were found to be highly prevalent, or the most frequent strains, in different regions of Argentina.
The molecular analysis, with high homogeneity in the G and P sequences, suggests the introduction of a G12[P8] strain in the local population, reaching sufficient fitness to spread through transmission among humans across the territory and become the main cause of the 2010–2011 rotavirus epidemic. The sequence of the VP7 segment in this strain could be differentiated from that of a G12 strain detected in Gipuzkoa in 2005, suggesting that the G12 strains sporadically detected in 2004 and 2005 were the result of previous introductions that failed to spread. The P-type [P8] is the most frequently associated with G12 in Europe, more frequently than [P6] and especially [P4] [Reference Iturriza-Gómara8]. Molecular studies have suggested that the novel G12[P8] strains could have been formed by the introduction of a VP7 gene into a globally common rotavirus strain [Reference Freeman11].
In conclusion, the results of this study indicate that rotavirus G12 should be considered as an emerging and clinically important G-type that could cause high-impact seasonal epidemics with similar characteristics to those caused by the major human rotavirus strains G1–G4 and G9. Surveillance of circulating rotavirus genotypes should be increased, especially in view of their importance in the development of effective rotavirus vaccines.
ACKNOWLEDGEMENTS
We thank the Pharmaceutical Union of Gipuzkoa for information on the number of rotavirus vaccine doses sold in Gipuzkoa in 2009 and 2010, which allowed us to estimate vaccination coverage. We are also grateful to EuroRotaNet for funding for genotyping of rotavirus (2009–2011).
DECLARATION OF INTEREST
None.





