Introduction
The genus Cruzia Travassos, 1917 comprises 10 species that parasitize the caecum and large intestine mainly of mammals but also amphibians and reptiles (Sprehn, Reference Sprehn1932; Ruiz, Reference Ruiz1947; Wolfgang, Reference Wolfgang1951; Crites, Reference Crites1956; Wahid, Reference Wahid1964; Costa, Reference Costa1965; Ubelaker and Younus, Reference Ubelaker and Younus1965; Guerrero, Reference Guerrero1971; Adnet et al., Reference Adnet, Anjos, Menezes-Oliveira and Lanfredi2009; Vieira et al., Reference Vieira, Gonçalves, Lima, Sousa and Muniz-Pereira2020). Up to now, 3 species of Cruzia have been reported infecting marsupials in the Americas: Cruzia cameroni Wolfgang, Reference Wolfgang1951 infecting Didelphis marsupialis Linnaeus, 1758 in Trinidad, Cruzia americana Maplestone, 1930 infecting D. marsupialis and Didelphis virginiana in the USA and Cruzia tentaculata Travassos, 1917 infecting Caluromys philander (Linnaeus, 1758), Didelphis albiventris Lund, 1841, Didelphis aurita Wied-Neuwied, 1826, D. marsupialis, Marmosa murina (Linnaeus, 1758), Metachirus myosuros (Temminck, 1824), Metachirus nudicaudatus (Geoffroy, 1803), Monodelphis domestica, Wagner, 1842 and Philander opossum Linnaeus, 1758, with the larger geographic distribution, occurring in Argentina, Bolivia, Brazil, Colombia, Paraguay, Peru, French Guiana and Mexico (Travassos, Reference Travassos1922; Ruiz, Reference Ruiz1947; Wolfgang, Reference Wolfgang1951; Crites, Reference Crites1956; Pinto and Gomes, Reference Pinto and Gomes1980; Martínez, Reference Martínez1986, Reference Martínez1987; Caneda-Guzman, Reference Caneda-Gusman1997; Gomes et al., Reference Gomes, Cruz, Vicente and Pinto2003; Adnet et al., Reference Adnet, Anjos, Menezes-Oliveira and Lanfredi2009; Tantaleán et al., Reference Tantaleán, Díaz, Sánchez and Portocarrero2010; Jiménez et al., Reference Jiménez, Catzeflis and Gardner2011; Mollericona and Nallar, Reference Mollericona and Nallar2014; Chero et al., Reference Chero, Sáez, Mendoza-Vidaurre, Iannacone and Cruces2017; Zabott et al., Reference Zabott, Pinto, de Marco Viott, Gruchouskei and de Barros Bittencourt2017; Li, Reference Li2019; Cirino et al., Reference Cirino, Costa, Maldonado and Gentile2020). Cruzia tentaculata is a generalist species with a monoxenous life cycle (Anderson, Reference Anderson2000), although the involvement of gastropod molluscan acting as intermediate hosts has been recently reported (Ramos-de-Souza et al., Reference Ramos-de-Souza, Maldonado, Vilela, Andrade-Silva, Barbosa, Gomes and Thiengo2021).
Didelphine marsupials have high abundance indices of parasitism by C. tentaculata in the Atlantic Forest, which suggests that they play an important role in the maintenance of the parasite metapopulation (Gardner, Reference Gardner1970; Cerqueira, Reference Cerqueira1985; Cáceres, Reference Cáceres2012; Chemisquy and Flores, Reference Chemisquy and Flores2012; Paglia et al., Reference Paglia, da Fonseca, Rylands, Herrmann, Aguiar, Chiarello, Leite, Costa, Siciliano, Kierulff, Mendes, Tavares, Mittermeier and Patton2012; Cole and Viney, Reference Cole and Viney2018; Voss et al., Reference Voss, Díaz-Nieto and Jansa2018). Some processes influencing the population structure of parasites include host mobility, geographical distance between different populations, gene flow and strength of genetic drift (Cole and Viney, Reference Cole and Viney2018). In addition to population genetic structure, parasite geographical variation can also be assessed at the phenotypic level by means of morphological and morphometric studies.
It has been suggested that the use of several host species allows nematodes to cross or colonize a wider range of habitats than would be possible using only a single host, and this tends to promote the genetic flow among parasite populations (Cole and Viney, Reference Cole and Viney2018). The aim was to investigate the existence of cryptic species within C. tentaculata using the null hypothesis that C. tentaculata is a single, panmictic population.
We hypothesized that (i) C. tentaculata represents a panmictic population along the Atlantic Forest, infecting several species of marsupials of the subfamily Didelphinae (host generalist), suggested by the fact that D. aurita and D. albiventris have large vagility compared to other small mammals, with the ability to cross degraded areas, exploring anthropized rural and urban environments; and (ii) C. tentaculata specimens present morphological variation related to the host localities.
Materials and methods
Study areas and sample collection
Specimens of C. tentaculata were recovered from 3 species of marsupials of the subfamily Didelphinae (D. aurita, D. albiventris and P. quica). These 3 species are the most commonly found and easily captured along the Atlantic Forest on the east coast of South America. Nine different localities were sampled along the Atlantic Forest biome: the municipality of Santa Rita (SRT-PB) in the state of Paraiba, the municipalities of São Cristovão (SCR-SE) and Capela (CAP-SE) in the state of Sergipe, the municipalities of Petrópolis (PET-RJ) and Rio de Janeiro (RIO-RJ) in the state of Rio de Janeiro, the municipality of Conceição dos Ouros (CDO-MG) in the state of Minas Gerais, the municipalities of Curitiba (CUR-PR) and Ponta Grossa (PGR-PR) in the state of Paraná and the municipality of Porto Alegre (POA-RS) in the state of Rio Grande do Sul. The number of helminths studied for each locality was based on the number of hosts collected in each locality and on the parasitic load of each animal to obtain representative samples from each local population (Table 1, Fig. 1).

Fig. 1. Identification of Cruzia tentaculata locality and host species collections carried out along the Atlantic Forest (shaded area in the map).
Table 1. Geographic locality, abbreviation, number of hosts, host species, number of specimens of Cruzia tentaculata used for genetic and morphometric studies separated by slash and geographic coordinates

Marsupials were captured using Tomahawk Live Trap (Hazelhurst, Wisconsin) model 201 traps (16″ × 5″ × 5″). Each trap was baited with a generalist mixture of peanut butter, banana, oats and bacon (nut, fruit, grain and animal protein), preferring odorous items for mammals' attraction. After capture, the marsupials were conducted to a field laboratory, where they were anaesthetized, killed and had their bionomic data and samples taken for different studies.
The marsupials were necropsied, and their organs were removed and placed separately in Petri dishes, washed in saline solution (0.9% NaCl) and dissected with the aid of a stereoscopic microscope. Specimens of C. tentaculata were recovered through examination of the host caecum and large intestine. The helminths were washed in saline solution and stored in 70% ethanol for further morphological, morphometric and molecular studies.
For morphological characterization and morphometric description, the specimens were clarified in 25% glycerinated alcohol, mounted on a slide under a coverslip, identified and measured with the aid of camera lucida attached to a Nikon Eclipse E200 MV R microscope. The identification of species by morphology and morphometry was performed using articles describing species of the genus Cruzia and taxonomic keys according to Vieira et al. (Reference Vieira, Gonçalves, Lima, Sousa and Muniz-Pereira2020). The morphometric characteristics analysed for males and females were the length and width of the oral capsule, length and width of the body, length and width of the oesophagus, length and width of the bulb, distance from the nerve ring and excretory pore with respect to the anterior extremity and length of the intestinal diverticulum. Only for males were the following characteristics measured: length of the spicules, length of the gubernaculum and distance from the cloaca to the posterior extremity. For females, the following characteristics were also measured: the distance from the vulva to the anterior extremity and from the anus to the posterior extremity. The specimens used in the morphometric analyses were not the same as those used in the molecular analyses and in the morphological analyses by scanning electron microscopy (SEM), so it was not possible to correlate them. For SEM analyses, the fixed nematodes were washed in 70% ethanol, dehydrated in a graded ethanol series (70–100%), critical point, dried in CO2, free-hand fractured, mounted on metallic stubs and coated with gold. The samples were analysed using a scanning electron microscope Jeol JSM 6390LV on the Electron Microscopy Platform Rudolf Barth, Instituto Oswaldo Cruz, Fiocruz.
Discriminant analysis of principal components
A discriminant analysis of principal components (DAPC) (Jombart et al., Reference Jombart, Devillard and Balloux2010) was performed to compare C. tentaculata specimen measurements among localities (PET-RJ, RIO-RJ, CUR-PR, PGR-PR, POA-RS, SCR-SE, CAP-SE, CDO-MG) and among host species (D. albiventris, D. aurita and P. quica). In this analysis, the same morphometric characteristics described above were used. The DAPC describes variations between defined groups, selecting principal components (PCs) that explain the greatest variation between groups while minimizing the variation within each group (Jombart and Collins, Reference Jombart and Collins2015). To identify the optimal number of PCs to be retained by DAPC and select the components associated with the lowest root mean-squared error, we used cross-validation optimization (Jombart and Collins, Reference Jombart and Collins2015). We determined the percentage of C. tentaculata specimens correctly classified within their original group. Finally, we tested the significance of the differences found within each group using Wilks' Lambda statistic.
DAPC analysis was performed for both male and female specimens of C. tentaculata, using only those specimens with sufficient information on morphometric measurements within each category among the groups represented by locality and host species. For male specimens, the locality of CUR-PR was removed due to its small sample size and the absence of morphometric measurements resulting from the preservation status of the material. Similarly, for female specimens, the locality of PGR-PR, POA-RS and SRT-PB and the species P. quica were removed, also due to small sample size or due to poor preservation of the material, preventing the visualizations of some structures. DAPC was performed using the package ‘adegenet’ (Jombart, Reference Jombart2008), and one-way MANOVA – Wilks' lambda was performed using the package ‘rrcov’ (Todorov and Filzmoser, Reference Todorov and Filzmoser2009), both in the R software environment version 4.1.2 (R Core Team, 2021).
Molecular and phylogenetic analyses
For molecular characterization, total genomic DNA of C. tentaculata specimens was isolated using the extraction kit NucleoSpin Tissue (Macherey-Nagel, Düren, Germany) according to the manufacturer's instructions and stored at −20°C until use. Partial cytochrome c oxidase subunit 1 (MT-CO1) mitochondrial gene fragments were amplified by polymerase chain reaction (PCR) using the primer cocktail described by Prosser et al. (Reference Prosser, Velarde-Aguilar, León-Règagnon and Hebert2013). Amplifications were performed at a final volume of 25 μL for each sample, comprising 12.5 μL of PCR Master Mix (Taq DNA polymerase 50 units mL−1, dATP 400 μ m, dGTP 400 μ m, dCTP 400 μ m, dTTP 400 μ m, MgCl2 3 mm) (Promega, Madison, Wisconsin, United States); 10.5 μL of ultrapure water for molecular biology, 0.5 μL of sense (primers Nem_F1, Nem_F2 and Nem_F3) and of anti-sense (primers Nem_R1, Nem_R2 and Nem_R3) oligonucleotide cocktail (10 μ m); and 1.0 μL of genomic DNA (~1–4 ng μL−1). We used a Veriti Thermal Cycler (Life Technologies) thermocycler under the following thermal cycling conditions: initial denaturation at 94°C for 1 min; 5 cycles of denaturation at 94°C for 40 s, hybridization at 45°C for 40 s, and extension at 72°C for 1 min; followed by 35 cycles of denaturation at 94°C for 40 s, hybridization at 51°C for 40 s, and extension at 72°C for 1 min; and a final extension at 72°C for 5 min (Prosser et al., Reference Prosser, Velarde-Aguilar, León-Règagnon and Hebert2013).
The resulting amplicons were visualized using 1.5% agarose gel electrophoresis with GelRed Nucleic Acid Stain (Biotium, Hayward, California, USA) on an ultraviolet light transilluminator. Successfully amplified PCR products were purified using the Illustra GFX PCR DNA and Gel Band kit (GE Healthcare, Chicago, Illinois, USA) according to the manufacturer's instructions and sequenced using the BigDye Terminator v3.1 Cycle Sequencing kit (Applied Biosystems, USA). The reactions were prepared at a final volume of 10 μL for each sample using the primer cocktail described by Prosser et al. (Reference Prosser, Velarde-Aguilar, León-Règagnon and Hebert2013). Sequencing reactions were performed on a Veriti Thermal Cycle (Life Technologies) thermocycler under the following cycling conditions: denaturation at 94°C for 10 s, hybridization at 50°C for 5 s and extension at 60°C for 4 min, followed by 39 repetitions of this cycle. Subsequently, cycle-sequencing product precipitation, formamide resuspension and analysis using the ABI3730xl DNA Analyzer (Applied Biosystems) were conducted at the Capillary Sequencing (SANGER) Platform of the Fiocruz Technological Platforms Network at the Oswaldo Cruz Institute (https://plataformas.fiocruz.br/unidades/RPT01A).
After sequencing, the forward and reverse sequences obtained were assembled into contigs and edited for ambiguities using the Geneious R9 software package (Kearse et al., Reference Kearse, Moir, Wilson, Stones-Havas, Cheun, Sturrock, Buxton, Cooper, Markowitz, Duran, Thierer, Ashton, Meintjes and Drummond2012). To build our data set, we include sequences of species representatives of the order Ascaridida Skrjabin & Shulz, 1940. As an outgroup, we added sequences of Ascaris lumbricoides (Linnaeus, 1758); Baylisascaris schroederi (Mclntosh, 1939); Parascaris equorum (Goeze, 1782); Pseudoterranova krabbei Paggi, Mattiucci, Gibson, Berland, Nascetti, Cianchi and Bullini, 2000; Toxascaris leonina (Von Linstow, 1902); and Toxocara cati (Schrank, 1788) based on phylogenetic proximity to the Kathlaniidae family (Table 2). Then, they were aligned by multiple alignment using the MUSCLE algorithms (Edgar, Reference Edgar2004) on the Translator X server (Abascal et al., Reference Abascal, Zardoya and Telford2010), which employs amino acid translations to align protein-coding nucleotide sequences.
Table 2. Species, GenBank accession number, geographic locality and host of Ascaridida GenBank sequences used in this study

Phylogenetic analyses were performed using the maximum likelihood (ML) and Bayesian inference (BI) methods. Phylogenetic reconstructions by ML were conducted using the online web server PhyML 3.0 (Guindon et al., Reference Guindon, Dufayard, Lefort, Anisimova, Hordijk and Gascuel2010). The selection of the nucleotide substitution model was performed with SMS (Smart Model Selection) (Lefort et al., Reference Lefort, Longueville and Gascuel2017) in PhyML using the Akaike information criterion (AIC). The nucleotide replacement model selected by the AIC in PhyML SMS was GTR + R. Statistical support to evaluate the reliability of the branches was obtained by bootstrap (BP) with 1000 pseudoreplicas and the approximate likelihood-ratio test for branches (aLRT). BI analyses were conducted using the program MrBayes 3.2.6 (Ronquist et al., Reference Ronquist, Meslenko, Mark, Ayres, Darling, Höhna, Larget, Liu, Suchard and Huelsenbeck2012), with independent GTR + I + G nucleotide substitution models for each codon position and unlinking base frequencies and parameters. Sampling was performed every 100 generations for 10 000 000 generations, with 4 simultaneous chains, in 2 runs, by MCMC. The robustness of the nodes was evaluated by Bayesian posterior probabilities (BPP), calculated after the removal of a burn-in fraction of 25%. To evaluate the adequacy of our sampling, we used the program Tracer 1.6 (Rambaut et al., Reference Rambaut, Suchard, Xie and Drummond2014) to calculate the effective sample size (ESS) of each parameter. MrBayes was run on XSEDE using the CIPRES Science Gateway (Miller et al., Reference Miller, Pfeiffer and Schwartz2010).
Population genetic analyses
To determine whether the genes were under selection, we performed neutrality tests to identify whether there was a deviation of the balance. Tajima's D (Tajima, Reference Tajima1989) and Fu's Fs (Fu, Reference Fu1997) neutrality tests were performed for each of the previously determined groups (i.e. based on hosts and locality) in the program Arlequin 3.5.2.2 (Excoffier and Lischer, Reference Excoffier and Lischer2010). To identify population structuring, fixation indices (Fst) (Wright, Reference Wright1978) were calculated to evaluate genetic differentiation between localities. Molecular variance analysis (AMOVA) (Excoffier et al., Reference Excoffier, Smouse and Quattro1992) was used to analyse the genetic variability between and within groups previously defined using the program Arlequin, and we also performed a Bayesian clustering analysis implemented in the program Bayesian Analysis of Population Structure (BAPS). Haplotype networks were calculated for the C. tentaculata sequences using the median joining (MJ) method (Bandelt et al., Reference Bandelt, Forster and Röhl1999) in the program PopART 1.7 (Leigh and Bryant, Reference Leigh and Bryant2015) to assess the distribution and sharing of haplotypes among populations.
The correlation between the genetic distances of all specimens of C. tentaculata and the geographical distances, considering all the localities studied, was investigated using the Mantel correlation test (Legendre and Legendre, Reference Legendre and Legendre2012). A genetic distance matrix was calculated in the ‘ape’ package (Paradis and Schliep, Reference Paradis and Schliep2019), the geographic distance matrix was calculated in the ‘fields’ package (Nychka et al., Reference Nychka, Furrer, Paige and Sain2017), and the Mantel correlation test was performed in the ‘vegan’ package (Oksanen et al., Reference Oksanen, Blanchet, Friendly, Kindt, Legendre, McGlinn, Minchin, O'Hara, Simpson, Solymos, Stevens, Szoecs and Wagner2020), all using the software environment R 4.1.0 (R Core Team, 2021).
Results
Morphological analysis by light, scanning electron microscopy and morphometric analysis
The measured male specimens of C. tentaculata have a body length ranging from 8.52 to 13.63 mm and females ranging from 10.52 to 16.13 mm. The anterior opening of the nematodes in both sexes is surrounded by 3 subtriangular lips, a dorsal lip with a pair of papillae on each side and 2 latero-ventral lips with a papilla and amphids on each side. Lips have 2 small teeth located on the inner margin. The pharynx has 3 rows each with 2 faces provided of 12 longitudinal pairs of cuticular lamellae and a tricuspid valve at the base (Fig. 2A). The oesophagus is cylindrical and long, divided into muscle and glandular portions, presenting a prebulbar dilation followed by a well-developed bulb containing 1 valve and accompanied by bulb valves at the base. The nerve ring is in the anterior oesophageal region, and the excretory pore is in the posterior third of the oesophageal region. The intestinal diverticulum is directed to the anterior region, exceeding the threshold of the oesophageal bulb. Males were curved ventrally in the posterior region, and the cloaca opening was at the caudal end. The caudal ventral region shows 10 pairs of bud-like papillae, 3 precloacal pairs, 3 ad-cloacal pairs and 4 postcloacal pairs, as well as a single papilla at the anterior edge of the cloacal opening. The spicules are subequal and alates, and the gubernaculum is triangular and has a slender caudal alae (Fig. 2B and C). The females are didelphic and amphidelphic, with the vulva in the preequatorial region and the tail ending in a thin tip.

Fig. 2. Light microscopy photographs of an adult male specimen of Cruzia tentaculata from the SRT-PB locality. (A) Details of the anterior region showing the lips (Lp), cuticular lamellae (Cl), tricuspid valve (Tv) and muscular part of the esophagus (Es) (bar: 50 μm). (B) Ventral view of the posterior region of the male showing spicules (Sp), gubernaculum (Gb), small caudal wing (Cw), 2 pairs of precloacal papillae (Pc), 3 pairs of ad-cloacal papillae (Acp) and 1 postcloacal papilla (Pcp) (bar: 50 μm). (C) Lateral view of the posterior region of the male showing spicules (Sp), gubernaculum (Gb), cloaca (Cl) and 1 postcloacal papilla (Pcp) (bar: 100 μm).
SEM analyses were performed to observe the cuticular lamellae of C. tentaculata specimens. The preparation required a longitudinal fracture of the oral capsule of some specimens. We observed 3 rows with 2 longitudinal facets with 12 pairs of well-developed cuticular lamellae (Fig. 3A and B).

Fig. 3. Scanning electron microscopy image of the anterior region of adults of Cruzia tentaculata. A–D: anterior extremity with 3 lips with a pair of small teeth located on the inner margins (triple arrow), dorsal lips (Dl) and latero-ventral (Vl). E and F: Anterior region of the nematodes fractured longitudinally, showing the internal surface of the pharynx ornamented with a row of pharyngeal lamellae with 12 pairs of cuticular lamellae (small arrows).
Discriminant analysis of principal components
MANOVA of morphometric variables showed that female specimens of C. tentaculata were significantly different among localities and host species (Table 3); however, this difference was more pronounced for the former, given the lower Wilks' lambda value for localities (Wilks' lambda < 0.01; P = <0.01) in relation to host species (Wilks' lambda = 0.07; P = <0.01) (Table 3). Males were significantly different among localities (Wilks' lambda < 0.01; P < 0.01) and among hosts (Wilks' lambda = 0.02; P = 0.05), although to a lesser extent in the latter (Table 3).
Table 3. Morphometric divergence of Cruzia tentaculata male and female specimens

d.f., degrees of freedom.
Comparison between groups (host species and locality) based on values of the classical and robust one-way MANOVA – Wilks' lambda.
In the DAPC for female parasites, 4 principal components (PCs) were retained to discriminate host species groups, and 2 PCs were retained to discriminate locality groups. For males, 2 PCs were retained to discriminate both host species and locality groups. In each of these DAPCs, the principal components retained explained approximately > 98% of the total variance. The proportion of female specimens correctly classified to their original groups was approximately 50% for localities and 86% for host species. For males, the proportion of specimens correctly classified to their original groups was approximately 57% for localities and 75% for host species.
When analysing the differences between localities, DAPC identified 2 sets of localities for females, 1 from the southeast region (RIO-RJ, PET-RJ and CDO-MG) and the other formed by the localities of the northeast region (CAP-SE and SCR-SE) and (CUR-PR) southern region (Fig. 4A). For males, 2 sets of localities were formed, 1 formed by a locality of the northeast region (SCR-SE) and a locality of the southern region (PGR-PR) and another formed by a locality of the southeast region (RIO-RJ, PET-RJ and CDO-MG) and a locality of the southern region (POA-RS) (Fig. 4B). The locality CAP-SE was isolated in the DAPC plot (Fig. 4B), indicating that the male specimens studied in this locality had unique morphometric characteristics (e.g. higher mean values of morphometric variables or lower variation of these variables among specimens in this locality compared to the others). In turn, as pointed out by MANOVA, differences between host species in DAPC were less evident than between localities, since total isolation was not observed between parasites across host species (Fig. 5A and B), which may indicate a greater degree of morphometric similarity between specimens of C. tentaculata occurring in different host species than among some localities.
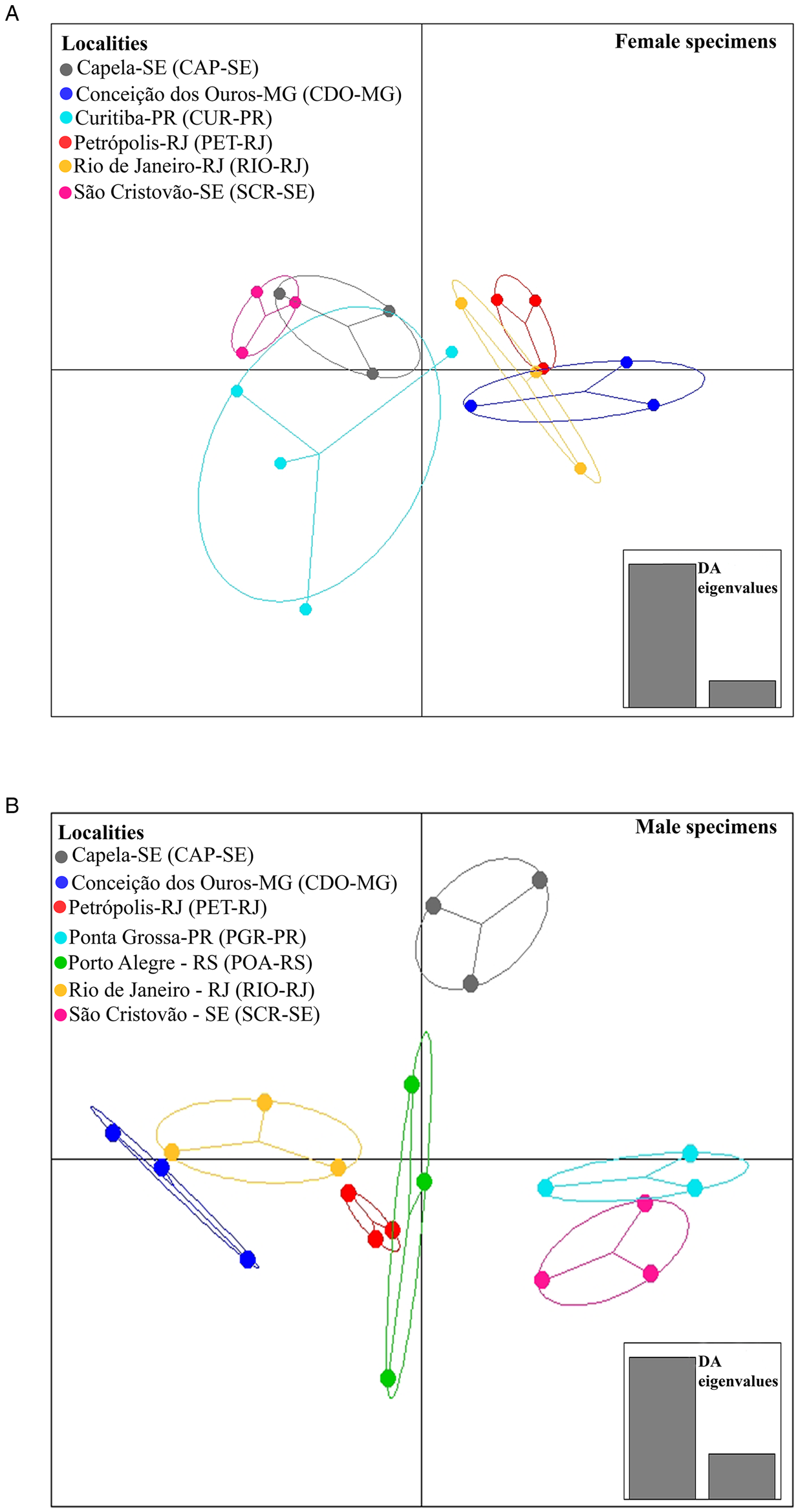
Fig. 4. Population clusters for female (A) and male (B) Cruzia tentaculata specimens, based on DAPC along with discriminant analysis (DA) eigenvalues, showing morphometric variations between localities.

Fig. 5. Population clusters for female (A) and male (B) Cruzia tentaculata specimens, based on DAPC along with discriminant analysis (DA) eigenvalues, showing morphometric variations between host species.
The variables that best discriminated parasites between the localities for female specimens were the length of the intestinal diverticulum and the body length (Fig. 6A). For males, those variables were the length of the intestinal diverticulum, the excretory pore and the length of the oesophagus (Fig. 6B). Regarding host species, the variables that best discriminated female parasites were body length, nerve ring, oesophagus length and vulva position (Fig. 7A). For males, those variables were the length of the intestinal diverticulum, the length of the excretory pore, the length of the spicules (short and long), and the body width (Fig. 7B).

Fig. 6. Morphometric variable contributions to Cruzia tentaculata specimens among localities. (A) Morphometric variable contributions to female Cruzia tentaculata specimens; (B) Morphometric variable contributions to male Cruzia tentaculata specimens.

Fig. 7. Morphometric variable contributions to Cruzia tentaculata specimens among host species. (A) Morphometric variable contributions to female Cruzia tentaculata specimens; (B) Morphometric variable contributions to male Cruzia tentaculata specimens.
Genetic diversity indices and population structure
In the present study, among 164 partial sequences of the MT-CO1 gene of C. tentaculata studied, 144 haplotypes with 161 polymorphic sites were found, and the mean nucleotide diversity and haplotypic diversity were 0.03480 and 0.998, respectively. To estimate the genetic diversity indices, the C. tentaculata sequences were organized into groups according to the host species and localities studied (Table 4).
Table 4. Cruzia tentaculata specimens partial MT-CO1 gene sequences genetic diversity based on host species and locality

N, sample size; H, number of haplotypes; S, number of polymorphic sites; Hd, haplotype diversity; π, nucleotide diversity.
Three matrices were constructed. The first was generated to calculate genetic diversity indices, AMOVA, Fst, Tajima's D and Fu's Fs neutrality tests, and the Mantel test, containing only the sequences of C. tentaculata generated in the present study, totalling 164 sequences with 693 bp. The second was composed of sequences obtained in the present study and 2 sequences of C. tentaculata species from GenBank, totalling 166 sequences with 693 bp, used to calculate haplotype networks and the Bayesian clustering analysis. The third matrix was composed of a representative of each haplotype obtained in the present study, 2 sequences of species of C. tentaculata from GenBank as an ingroup, and 6 sequences from GenBank of the order Ascaridida as an outgroup, resulting in 152 sequences, each with 693 base pairs (bp) used for phylogenetic inferences.
In the ML analysis, the nucleotide replacement model selected by the AIC in PhyML SMS was GTR + R, with an estimated invariable site proportion of 0.000, estimated gamma parameter of 1.000 and variation of rates with 4 categories. The most likely tree presented an LnL of −4970.750524. The estimated base frequencies were A = 0.15645, C = 0.06752, G = 0.26508, and T = 0.51095. For BI, the estimated mean marginal probability was −4982.4043, and the median was −4982.079. The ESSs for all parameters were above 100 samples and were effectively independent, indicating the robustness of our sampling. Phylogenetic reconstructions by ML and BI resulted in equal topologies. Specimens of C. tentaculata formed a monophyletic group in both phylogenies (aLRT = 0.99; BP-MV = 0.99; BPP = 1.00). In the ML (Supplementary material) and BI (Fig. 8), the group of C. tentaculata was composed of parasites from D. albiventris in the state of Paraíba (SRT), Sergipe (CAP and SCR), Paraná (CUR), Rio Grande do Sul (POA), by parasites from D. aurita in the states of Rio de Janeiro (RIO and PET), Paraná (CUR) and Minas Gerais (CDO) and from P. quica in the state of Paraná (PGR).

Fig. 8. Phylogenetic reconstruction by Bayesian inference (BI) based on partial MT-CO1 gene sequences (693 bp) of 164 specimens of Cruzia tentaculata, parasites of Didelphis albiventris (white), Didelphis aurita (black), and Philander quica (grey) from the Atlantic Forest biome, 2 sequences of species of Cruzia tentaculata from GenBank and 6 sequences of other species belonging to the order Ascaridida as an outgroup. Values at nodes are the Bayesian posterior probabilities (BPP). Colours represent each locality in relation to the network of haplotypes.
The haplotype network suggested separation of 2 groups: SRT-PB and SCR-SE (group I) and PET-RJ, RIO-RJ, POA-RS, CUR-PR, PGR-PR, SCR-SE, CAP-SE and CDO-MG (group II), separated by a genetic distance of 32 mutational steps. The sharing of haplotypes between localities and host species was observed. Haplotype H-5 was shared between the POA-RS and PET-RJ localities (Fig. 9). Haplotype H-5 was shared between D. albiventris and D. aurita. Interestingly, in the host haplotype network, there were 3 groups of haplotypes for D. albiventris at the periphery of the haplotypes of D. aurita (Fig. 10).

Fig. 9. Median-joining haplotype network based on partial MT-CO1 gene sequences (693 bp) of Cruzia tentaculata, a parasite of the caecum and large intestine of Didelphis albiventris, Didelphis aurita and Philander quica, from this study and 2 sequences of species of Cruzia tentaculata from GenBank. The size of the circles represents the frequency of haplotypes. The colours of the circles represent the occurrence locality of each haplotype. In the network, 2 groups were observed: group I (SRT-PB and SCR-SE) and group II (PET-RJ, RIO-RJ, POA-RS, CUR-PR, PGR-PR, SCR-SE, CAP-SE and CDO-MG).

Fig. 10. Median-joining haplotype network based on partial MT-CO1 gene sequences (693 bp) of Cruzia tentaculata, a parasite of the caecum and large intestine of Didelphis albiventris, Didelphis aurita and Philander quica, from this study and 2 sequences of species of Cruzia tentaculata from GenBank. The size of the circles represents the frequency of haplotypes. The colours of the circles represent the occurrence hosts of each haplotype.
Regarding hosts, the AMOVA indicated higher genetic variance within host (88.85%) species than between hosts. There was a higher genetic variation between localities (52.82%) than within localities (Table 5). The value of Fst for host species was significant when compared between D. aurita and D. albiventrtis (0. 002). For localities, the Fst values were significant between CDO-MG and SCR-SE, CDO-MG and CAP-SE, SCR-SE and POA-RS, SCR-SE and CUR-PR, SCR-SE and RIO-RJ, SCR-SE and CAP-SE, SCR-SE and SRT-PB, POA-RS and CUR-PR, POA-RS and CAP-SE, POA-RS and SRT-PB, CUR-PR and CAP-SE, CUR-PR and SRT-PB, RIO-RJ and CAP-SE and CAP-SE and SRT-PB (Table 6). The neutrality tests Tajima's D and Fu's Fs were determined for each group. For the comparisons formed by the host species, D values were significant only for the group of C. tentaculata parasites from D. aurita, while Fs values were significant for both D. aurita and D. albiventris. Considering the localities, the D values were not significant. The Fs values were significant for SRT-PB, PET-RJ, RIO-RJ, CUR-PR, CDO-MG and POA-RS (Table 7). According to the population structure using BAPS, specimens of C. tentaculata were distributed in 5 clusters (Fig. 11).

Fig. 11. Population structure of Cruzia tentaculata deduced by Bayesian analysis of population structure. Structural plot of 166 specimens of Cruzia tentaculata revealing 5 different major populations (clusters 1–5). The locations of each cluster are represented in the graph.
Table 5. Cruzia tentaculata specimens partial MT-CO1 gene sequence genetic variability by AMOVA based on host species and localities

d.f., degrees of freedom.
Table 6. Cruzia tentaculata specimens partial MT-CO1 gene sequences genetic variance estimation by Fst based on collection locality

Significant values are in bold (P < 0.05).
Table 7. Neutrality tests (Tajima's D and Fu's F's), with the respective P values, of Cruzia tentaculata specimens partial MT-CO1 gene sequences grouped into host species and locality

Significant values are in bold (P < 0.05).
The Mantel test indicated that approximately 68% of the genetic differentiation was explained by the spatial distance between the localities (correlation coefficient, r: 0.6764 P = 0.001) (Table 8, Fig. 12).

Fig. 12. Mantel test result showing the correlation between genetic and geographic distances in kilometres (r: 0.6764 P = 0.001).
Table 8. Geographic distance matrix in kilometres

Shared haplotype: H-5 (POA-RS and PET-RJ).
Discussion
Morphological diversity and morphometric comparison between Cruzia tentaculata
Morphological analyses performed by SEM and light microscopy confirmed the identification of the studied specimens as C. tentaculata, agreeing with the taxonomic key (Vieira et al., Reference Vieira, Gonçalves, Lima, Sousa and Muniz-Pereira2020) and articles discussing the species (Travassos, Reference Travassos1922; Adnet et al., Reference Adnet, Anjos, Menezes-Oliveira and Lanfredi2009). Most studies of morphological descriptions of the species do not examine the number of cuticular lamellae. According to Ruiz (Reference Ruiz1947) and Adnet et al. (Reference Adnet, Anjos, Menezes-Oliveira and Lanfredi2009), C. tentaculata presents only 10 pairs of tooth-like structures. Crites (Reference Crites1956) examined 6 male specimens of C. tentaculata parasitic in D. aurita collected in Brazil and reported that specimens of C. tentaculata have 8–14, generally 10, cuticular lamellae. Comparing our samples with 11 specimens of C. tentaculata from the Helminthological collection of the Oswaldo Cruz Institute (CHIOC) (Fiocruz-CHIOC 1556 – parasitic in Didelphis sp. identified by Travassos; Fiocruz-CHIOC 33678 – parasitic in Didelphis aurita identified by Vicente & Cruz, 1997; and Fiocruz-CHIOC 35624 – parasite of Didelphis aurita identified by Adnet et al., Reference Adnet, Anjos, Menezes-Oliveira and Lanfredi2009), all observed specimens presented 12 pairs of cuticular lamellae similar to those observed in the specimens of the present study. These findings corroborate Crites (Reference Crites1956), who suggested a range of 8–14 cuticular lamellae and should be considered in the identification key of the species according to Vieira et al. (Reference Vieira, Gonçalves, Lima, Sousa and Muniz-Pereira2020).
Li (Reference Li2019) analysed specimens of C. americana collected from the marsupial D. virginiana in the United States using light microscopy and scanning electron microscopy for the first time. The SEM images revealed the internal structures of the pharynx of C. americana and the presence of pharyngeal lamellae instead of pharyngeal teeth and/or teeth-like structures. In the present study, when analysing specimens of C. tentaculata by light microscopy and SEM, we observed that the structures in the pharynx are thin, flattened and parallel to each other. With this, we agree that these structures are actually cuticular lamellae, previously described as denticles or teeth-like structures.
Among the species of the genus Cruzia, only C. tentaculata C. cameroni and C. americana were reported parasitizing marsupials of the subfamily Didelphidae. Cruzia americana differs from C. tentaculata because it has 15–18 cuticular lamellae in each longitudinal line and 5 pairs of postcloacal papillae, while C. tentaculada has 4 pairs of postcloacal papillae. In addition, C. tentaculata has a single papilla on the anterior edge of the cloaca, but this papilla was not found in specimens of C. americana (Travassos, Reference Travassos1922; Ruiz, Reference Ruiz1947; Wolfgang, Reference Wolfgang1951; Crites, Reference Crites1956; Adnet et al., Reference Adnet, Anjos, Menezes-Oliveira and Lanfredi2009; Li, Reference Li2019). Li (Reference Li2019) also reported that the intestinal diverticulum of C. americana is distinctly longer than that of C. tentaculata. Cruzia cameroni can be differentiated from C. tentaculata because it has 13–17 cuticular lamellae and distinctly larger spicules and does not have a single papilla on the anterior edge of the cloaca. In addition, C. cameroni has the vulva located in the equatorial region of the body, while the vulva of C. tentaculata is located in the pre-equatorial region.
Regarding the discriminant analyses, significant differences in morphometric traits of C. tentaculata were observed in both males and females either among localities or among host species. We hypothesize that these differences are related to several factors related to the coevolutionary processes of host–parasite interactions. Differences in host body size or host metabolic rates would influence the size of some helminth traits because, for having an endoparasitic way of life, they must be adapted to the host characteristics. Differences among localities can be attributed to distinct local ecological factors, which in turn could influence the host characteristics, as well as the free-living stages of the helminth. Such a hypothesis could only be confirmed with a more comprehensive study encompassing all the host species in which this helminth has been found and including measurements of environmental variables and host attributes.
Population structure of Cruzia tentaculata related to locality and hosts
The AMOVA showed a similar percentage of genetic variation between and within locations (51.58% and 48.42%, respectively). This fact is corroborated by the low and non-significant Fst values between localities, indicating low genetic differentiation, low geographic structure and high gene flow of the parasite populations among the studied areas. A mixture was found between the clusters (1, 3, 4 and 5), corroborating the haplotype network, the phylogenetic analyses and the low Fst values. In addition, cluster 2 recovered in BAPS was congruent with group II found in the haplotype network. However, the heterogeneity observed between populations can be attributed to geographic distance, corroborated by the Mantel test, which indicated a significant and high (68%) correlation between geographic and genetic distance, suggesting isolation by distance. Most of the genetic divergence, shown by haplotype networks, is between the northern populations and the southern ones, separated by 1200 km.
The phylogenetic analyses and the haplotype network showed that specimens of C. tentaculata were not correlated with the host species. In addition, the host species were also observed to share haplotypes. The pattern found in phylogenies was corroborated in the haplotype network, and no structure was observed associated with the host species of C. tentaculata, with AMOVA indicating greater genetic variation within each host species (88.85%). In addition, the Fst value was very low and significant and was more evident between D. aurita and D. albiventris. Didelphine marsupials are phylogenetically close and can occur in sympatry in regions of ecotone between the Atlantic Forest and the Cerrado, showing overlap in their diets, which allows parasite sharing and therefore sharing of their haplotypes (Leite et al., Reference Leite, Costa and Stallings1996). In population genetic structures, the host is a key element among parasite populations that determines genetic variability and gene flow (McCoy et al., Reference McCoy, Boulinier, Tirard and Michalakis2003), and gene flow is dependent on host displacement and encounters; thus, parasites partially share the phylogeographic history of their host (Nieberding et al., Reference Nieberding, Morand, Libois and Michaux2004, Reference Nieberding, Libois, Douady, Morand and Michaux2005).
Furthermore, the generalist characteristic of C. tentaculata infecting a wide range of host species, such as C. philander, D. marsupialis, Marmosa murinae, Metachirus myosuros, M. nudicaudatus, Monodelphis domestica and Philander opossum, may contribute to the homogenization of populations. In addition, Nascimento et al. (Reference Nascimento, Campos, Fraga and Barros2018) demonstrated that D. albiventris forms structured populations along the northeast/south-southeast axis (Nascimento et al., Reference Nascimento, Campos, Fraga and Barros2018), coinciding with the results found for C. tentaculata.
López-Caballero et al. (Reference López-Caballero, Mata-López, García-Varela and De León2015) conducted a study to analyse the population structure of the helminth Oligacanthorhynchus microcephalus (Rudolphi, 1819) (phylum Acanthocephala) in Didelphis marsupialis, D. virginiana and Philander opossum in central and southeastern Mexico. Phylogenetic analyses demonstrated a similar pattern to that of C. tentaculata, in which the specimens of O. microcephalus were not correlated with host species or geographical locality.
The high haplotypic diversity found in C. tentaculata was also identified in Haemonchus contortus (Rudolph, 1803), a parasite of sheep and goats (Dey et al., Reference Dey, Zhang, Begum, Alim, Hu and Alam2019). In the present study, of the 164 sequences of the MT-CO1 gene of C. tentaculata, 144 haplotypes with 161 polymorphic sites were corroborated by H. contortus, of which 85 sequences of the mitochondrial gene nad4 resulted in 77 haplotypes with 115 polymorphic sites. In addition, H. contortus showed a low Fst value, and high gene flow was observed among the subpopulations, similar to that observed in the analyses of C. tentaculata.
Our results showed that there is a possibility of having 2 lineages, 1 from the southeast (D. aurita) and another from the northeast (D. albiventris). The haplotype network showed the separation of the 2 groups. We believe that the centre of origin was in D. aurita (southeast) and that secondary infections infected D. albiventris (northeast), as shown in the haplotype network.
Demographic and biogeographic history of Cruzia tentaculata
Historical events leave signatures in the DNA, and neutrality tests can infer the demographic history of populations. Significantly negative values in neutrality tests indicate that the population is in an expansion process, showing a large number of rare haplotypes. Significant positive values in neutrality tests indicate that the species has experienced a population bottleneck, that is, the lack of rare haplotypes due to a drastic reduction in population size (Hartl and Clark, Reference Hartl and Clark2010). In the neutrality tests of the different groups studied, some of them presented significant negative values. Considering Fu's Fs related to the hosts D. aurita and D. albiventris and for most of the locations, the results indicated the occurrence of an expansion process, although not corroborated by Tajima's D. However, the signature of population expansion was corroborated by the genetic diversity indices that were performed for the localities and host groups. High haplotypic diversity and low nucleotide diversity were found for the MT-CO1 gene; that is, although there were a large number of haplotypes, they differed from each other by a few nucleotide substitutions. This finding indicates a recent population expansion (Avise, Reference Avise2000).
Pôrto et al. (Reference Pôrto, Cabral and Tabarelli2004) carried out an integrated study encompassing water resources, socioeconomic aspects, fauna, flora and environmental education, particularly regarding the biodiversity of highland humid forests in the northeast Atlantic Forest (Paraíba e Pernambuco states), and reported the occurrence of endemic species as well as Amazonian species. The coastal forest in the Brazilian northeast has been identified as an important centre of endemism in South America, which is influenced by the Amazonian biota in the north (Prance, Reference Prance1982, Reference Prance1987) and the Atlantic Forest in southern and southeastern Brazil (Andrade-Lima, Reference Andrade-Lima1960, Reference Prance1982). The authors reported a hypothesis for the origin of the phytophysiognomy of highland humid forests associated with climatic variations that occurred during the Pleistocene, which allowed the Atlantic Forest to occupy the Caatinga domains, followed by interglacial periods, and the formation of Atlantic Forest islands in the Caatinga domains named ‘refuges of diversity’ (Andrade-Lima, Reference Andrade-Lima1982). In fact, our results corroborated this hypothesis since we observed a population isolation of C. tentaculata coinciding with the historical aspects suggested for the constitution of the northeastern Atlantic Forest.
The ‘Hypothesis of refuges’ predicts that during the glacial periods, the flora and fauna of forested areas became isolated in different forest fragments (refuges), differentiating and leading to speciation. When the glacial period ended, the forests expanded, and the species met again. The Atlantic Forest has 5 areas of great genetic diversity, which correspond to forest refuges (Carnaval and Moritz, Reference Carnaval and Moritz2008). They are (1) a great refuge from the north of Espírito Santo state to the northern coast of Bahia state; (2) a large refuge in the southeast region, including the south of Rio de Janeiro state, the northeast of São Paulo state and the southeast of Minas Gerais state; (3) a small area on the coast of Alagoas and Pernambuco states; (4) a small area in Chapada Diamantina, in the interior of Bahia state; and (5) a small area in northeastern Mato Grosso do Sul state. According to Carnaval and Moritz (Reference Carnaval and Moritz2008), in non-refugee areas, molecular signatures of recent expansion are expected to be found. In our analyses, the groups recovered in the haplotype networks were distributed in regions that did not correspond to refuges, which suggests the expansion of 2 lines of C. tentaculata isolated from the forest areas of refuges that expanded to other regions during interglacial periods. These findings suggest that 1 of these lineages of C. tentaculata may have migrated from the northeast (Sergipe and Paraíba states), giving rise to group I. The second lineage may have differentiated and expanded by the northeast region (Sergipe state), southeast (Rio de Janeiro and Minas Gerais states) and dispersed to the south (Paraná and Rio Grande do Sul states), giving rise to group II.
Conclusions/future directions
Some analyses, such as the Mantel test, AMOVA, the low values of Fst and the sharing of haplotypes, indicate that there is a high gene flow between the subpopulations of the Atlantic Forest, corroborating our initial hypothesis that the specimens of C. tentaculata may represent a panmictic population along the Atlantic Forest. However, we cannot affirm that the analysed individuals of C. tentaculata form a panmictic population, which is supported by the neutrality tests, morphometric differences found, and the haplotype networks, which showed the existence of 2 groups. Thus, despite the low genetic differentiation observed, the first hypothesis cannot be confirmed with the data of the present study, unlike the second hypothesis, which was corroborated. We suggest that the morphological differences and the haplotypes grouping into 2 groups observed are attributed to ecological differences, which can be explained by the hypothesis of refuges. We suggest a more comprehensive study including other molecular markers to corroborate the observed patterns and the inclusion of parasite samples from the Cerrado and Amazon biomes to test the hypothesis of refuges.
Supplementary material
The supplementary material for this article can be found at https://doi.org/10.1017/S0031182022000981.
Acknowledgements
We would like to thank the staff and students of Laboratório de Biologia e Parasitologia de Mamíferos Silvestres Reservatório at Fundação Oswaldo Cruz. Special thanks to Dr Paulo D'Andrea for the ICMBIO licence. We thank Dra. Liliani Marilia Tiepolo (UFPR) for providing samples of Ponta Grossa (Project CNPq/ICMBio/FAPs nº 18/2017; Process 421292/2017-2) and for the DNA Sequencing Platform at the Oswaldo Cruz Institute (PDTIS/FIOCRUZ). We are grateful to Rudolf Barth for the electron microscopy and Fernando de Oliveira dos Santos for helping with the map.
Author contributions
R. S., A. M. J. and R. V. V. co-designed the study. P. C.-E., R. G., R. M., R. V. V. and S. F. C.-N. conducted the biological material collections. E. J. L.-T. performed the scanning electron microscopy of the parasites. R. S. and A. M. J. identified and measured the parasites. R. S. performed all molecular biology laboratory procedures. T. S. C. conducted DAPC, MANOVA and Mantel tests. R. S., R. V. V. and K. V. conducted all the genetic analyses. R. S. drafted the manuscript and A. M. J., R. G. and R. V. V. revised it. All authors have read and approved the final manuscript.
Financial support
This project was financially supported by Conselho Nacional de Desenvolvimento Científico e Tecnológico – CNPq – PPBio Rede BioMA (457524/2012-0), Fundação Carlos Chagas Filho de Amparo à Pesquisa do Estado do Rio de Janeiro – FAPERJ (E-26/010.001597/2019), Instituto Oswaldo Cruz (IOC – FIOCRUZ), Serviço de Referência em Saúde at Fundação Oswaldo Cruz and Programa de Pós-Graduação em Biodiversidade e Saúde (IOC-FIOCRUZ). R. S. received grants from Instituto Oswaldo Cruz and from Coordenação de Aperfeiçoamento de Pessoal de Nível Superior Brazil (CAPES) – Finance Code 001; CNPq (R. G., 304355/2018-6), (S. F. C.-N., 164846/2020-4), (R. M., 313963/2018-5); and FAPERJ (T. S. C., E-26/204.421/2021), (R. M., E-26/203.274/2017, E-26/210.254/2018, E-26/200.967/2021).
Conflict of interest
The authors declare there are no conflicts of interest.
Ethical standards
The euthanasia of marsupials followed the guidelines of the Committee on The Care and Use of Animals of the American Mammal Society (Sikes, 2016) and the Brazilian Guide to Good Practices for Euthanasia in Animals by the Committee on Ethics, Bioethics and Animal Welfare of the Federal Council of Veterinary Medicine (CFMV, 2013). Authorizations for the capture and handling of mammals were issued by the Chico Mendes Institute for Biodiversity Conservation (ICMBio/SISBIO, licence numbers 13373-1, 45839-1 and 63023-4) and the State Institute of Environment of Rio de Janeiro (INEA, licence number 020/2011). The procedures were approved by the Animal Ethics Committees of the Oswaldo Cruz Foundation and the Oswaldo Cruz Institute (CEUA-FIOCRUZ, licence number LW-39/14, and CEUA-COI, licence number L-036/2018).
























