INTRODUCTION
Shigellosis remains a significant public health problem in developing countries where it is a major cause of diarrhoea-related morbidity and mortality, especially in children aged <5 years [Reference Ram1]. Shigella flexneri and Shigella sonnei have been reported as the most common causes of shigellosis in the developing world; in developed countries S. sonnei dominates [Reference Kotloff2, Reference Niyogi3]. Shigella dysenteriae and Shigella boydii are much less commonly reported as causes of shigellosis [Reference Ram1, Reference Kotloff2]. However, S. dysenteriae can be the cause of large outbreaks of diarrhoeal disease with severe consequences; the increased virulence is primarily due to the secretion of the phage-encoded Shiga toxin [Reference Acheson, Donohue-Rolfe, Keusch, Alouf and Freer4, Reference Kernéis5].
Several epidemiological studies have examined the burden of shigellosis in Egypt [Reference Putnam6–Reference Wierzba10]. Four of these studies examined the Shigella serotypes responsible for disease [Reference Putnam6–Reference Wasfy8, Reference Wierzba10]. The criteria used to enrol cases in each of these studies were different. Two studies used active surveillance of a birth cohort in which children aged <24 months were enrolled and remained in the study until aged 36 months [Reference Putnam6, Reference Abu-Elyazeed7], another study used passive surveillance of paediatric severe diarrhoea where every fifth child (0–59 months) was enrolled from two separate outpatient clinics [Reference Wierzba10], while a fourth study used passive surveillance within all admissions to a Cairo fever hospital (median age 14·1 years; range 4·8–23·4 years [Reference Wasfy8]. Despite the variable enrolment criteria, S. flexneri was the dominant species in all age groups in all four studies. However, in three of the four studies, S. dysenteriae was the second most common cause of shigellosis, followed by S. sonnei. S. boydii was least often associated with gastroenteritis.
Strain characterization of Egyptian isolates of Shigella beyond species type is uncommon and where data are available, it pertains almost exclusively to characterization of isolates of S. flexneri [Reference Ahmed11, Reference El-Gendy12]. Wasfy et al. [Reference Wasfy8] reported that the isolates of S. dysenteriae identified in all-age severe cases of diarrhoea were mainly subtypes 1, 2 or 3, but the percentages of isolates were not stated. No information has been reported for S. boydii.
Antibiotic treatment of shigellosis is increasingly complicated due to the rising levels of resistance to most widely available and inexpensive antibiotics such as ampicillin (AM), chloramphenicol (C), trimethoprim/sulfamethoxazole (SXT), and tetracycline (TE) [Reference Kotloff2, Reference Sack, Lyke, McLaughlin, Sack, Lyke and McLaughlin13]. In Egypt only a few studies have examined antimicrobial sensitivities to Shigella spp. [Reference Putnam6, Reference Wasfy8] and one study explored resistance in S. flexneri isolates [Reference Ahmed11]. Almost no data regarding subserotype, mechanism of antimicrobial resistance, and genetic diversity of S. dysenteriae and S. boydii have been published from Egypt, despite the importance of these pathogens as causes of paediatric diarrhoea.
Our overall goal was to integrate clinical and epidemiological data gathered regarding Shigella-associated diarrhoea in Egyptian children with advanced characterization of specific Shigella spp. In addition, we wished to document the basis for antibiotic resistance, with a particular emphasis on β-lactam and SXT antibiotics; commonly used to treat diarrhoea in the local paediatric population. In our first report [Reference Ahmed11] we presented data from children infected with S. flexneri. In this report we present data from children infected by S. dysenteriae and S. boydii exclusively.
METHODS
Bacterial strains and culture conditions
A total of 40 S. dysenteriae and 30 S. boydii isolates were analysed from stool samples collected from 70 children aged <5 years participating in paediatric cohort and paediatric severe diarrhoea studies conducted during the period 1998–2007, in rural Abu-Homos, Bahira Governorate, the periurban Fayoum district, or the urban district of Moqqatum Hills, Cairo. Briefly, three birth cohort studies centered in the Abu Homos field site were conducted from 1998 to 2007. Cohort study 1, conducted between 1998 and 2000, enrolled 462 children aged <1 month, with follow-up between 6 and 18 months; this study aimed to measure the efficacy of an enterotoxigenic Escherichia coli (ETEC) vaccine trial. Cohort study 2 was conducted to determine the age-specific incidence of Helicobacter pylori infection in infants and children (1998–2003); 277 children were enrolled within their first month of life and put under active surveillance until achieving their third birthday. Cohort study 3 (2004–2007) was conducted to determine the natural history of ETEC infection; 348 children aged <1 month were enrolled and followed for 2 years. Study subjects in all cohorts were visited twice weekly in their homes to determine if they had loose or liquid stools since the last visit. If such symptoms were present, a rectal swab and stool sample were collected and sent to NAMRU-3 for analysis. For childen enrolled in the paediatric severe diarrhoea studies at Abu Homos Hospital during May 2000 to September 2007, every fifth child aged <5 years who attended the paediatric clinic complaining of diarrhoea was approached for enrolment; a total of 3813 children were screened and enrolled. A total of 3457 children aged <5 years enrolled in outpatient hospital clinics at Moqqatum Hills during October 2002 to September 2007, and in Fayoum district (a hospital paediatric outpatient clinic) during August 2003 to September 2003 a total of 356 children were enrolled. In hospital studies, children aged <5 years who complained of diarrhoea had stool samples collected and rectal swabs performed, after informed consent, which were sent to NAMRU-3 for analysis.
In instances when Shigella was isolated more than once from the same child during the same diarrhoeal episode, only one isolate was examined. Rectal swabs were cultured on MacConkey agar (Becton Dickinson, USA) and Salmonella-Shigella agar (Becton Dickinson), and Shigella isolates were identified by standard microbiological procedures [Reference Bopp, Murray, Baron, Pfaller, Tenover and Yolken14]. Shigella isolates were frozen in trypticase soy broth supplemented with 15% glycerol at −70°C until required. Details of these studies have been described elsewhere [Reference Wierzba10, Reference Rao15].
Statistical analysis
The values of variables were counted and summarized in tables of frequency. The χ2 test was used for the analysis of categorical variables. When the frequency in a cell was lower than five, Fisher's exact test was used instead of χ2. Collected demographic and clinical data of all S. dysenteriae and S. boydii were analysed using SPSS version 15 (SPSS Inc., USA).
Serotyping
Serogroups were determined using commercial polyvalent group sera and visual inspection of slide agglutination assays as described by the manufacturer (Denka Seiken Co., Japan). Similarly, species serotypes were determined using either 12- or 18-monovalent sera specific for S. dysenteriae or S. boydii, respectively.
Antimicrobial susceptibility testing (AST)
Isolates of Shigella were characterized for antimicrobial susceptibility to seven antimicrobial compounds using the disk diffusion method according to the Clinical and Laboratory Standards Institute (CLSI) guidelines [16]. The antibiotic test panel included 10 μg AM; 30 μg C; 30 μg nalidixic acid (NA); 5 μg ciprofloxacin (CIP); 30 μg TE; SXT (1·25 μg trimethoprim/23·75 μg sulfamethoxazole); and 30 μg cephalothin (CR),. After incubation on Mueller–Hinton agar plates at 37°C for 18 h, zones of inhibition diameters were recorded. Results were interpreted according to CLSI guidelines as susceptible (S), intermediate (I) or resistant (R). For the purpose of reporting, we refer to both intermediate and susceptible isolates as ‘non-resistant’. Multidrug resistance (MDR) was defined as resistance to at least three classes of antibiotic as defined previously [Reference Putnam6]. Escherichia coli ATCC 25922, Staphylococcus aureus ATCC 25923 and Pseudomonas aeruginosa ATCC 27853 were used as control strains for AST according to CLSI guidelines [Reference Houng, Sethabutr and Echeverria17].
DNA isolation and PCR
Genomic DNA from bacterial isolates was prepared from whole-cell lysates as described previously [Reference Houng, Sethabutr and Echeverria17]. Isolates phenotypically resistant to AM and SXT were examined using oligonucleotides specific for bla TEM, bla OXA, class 1 and 2 integrons and sulfonamides genes (sulI and sulII). PCR amplicons were loaded into 2% agarose (Sigma-Aldrich Inc., USA) gels and DNA subjected to electrophoresis. Molecular mass of DNA fragments was determined by comparison with a commercial DNA standard (100-bp DNA ladder, Promega, USA). E. coli ATCC 25922, an antibiotic susceptible strain, was used as negative control in each PCR experiment.
The presence of class I integrons was inferred by amplification of a 280-bp fragment of the integrase gene intI1 using primers int1-F (5′-cctcccgcacgatgatc-) and intI1-R (5′-tccacgcatcgtcaggc-) as described previously [Reference Goldstein18]. Class II integrons were inferred by amplification of a 233-bp segment of the integrase gene intI2 using primers intI2-F (5′-ttattgctgggattaggc-) and intI2-R (5′-acggctaccctctgttatc-) [Reference Goldstein18]. Detection of resistance to β-lactams was performed amplifying 643- and 599-bp fragments using the following primer sets: bla TEM-F (5′-cagcggtaagatccttgaga-) and bla TEM-R (5′-actccccgtcgtgtagataa-), and bla OXA1-F (5′-aatggcaccagattcaactt-) and bla OXA1-R (5′-cttggcttttatgcttgatg-) as described previously [Reference Chen19]. Primers directed towards internal portions of the sulI and sulII genes amplified fragments of 433 bp (sulI-F, 5′-cggcgtg ggctacctgaacg-; sulI-R, 5′-gccgatcgcgtgaagttccg-) and 293 bp (sulII-F, 5′-gcgctcaaggcagatggcatt-; sul1II-R, 5′-gcgtttgataccggcacccgt), respectively [Reference Kerrn20].
XbaI pulsed-field gel electrophoresis (XbaI–PFGE) and data analysis
XbaI digestion and subsequent PFGE of chromosomal DNA was used to detect genetic diversity among S. dysenteriae and S. boydii isolates. Preparation of intact genomic DNA, digestion and electrophoresis conditions were as described previously [Reference Ribot21]. XbaI–PFGE profiles were compared by automated analysis using the BioNumerics Software package (version 5.10; Applied Maths, USA) and confirmed by visual inspection. Similarity between PFGE banding patterns was calculated using the Dice coefficient with a 2% tolerance for the band migration distance and clustering was performed using the complete linkage method; this analysis method is considered a more strict interpretation of banding patterns than UPGMA [Reference Vauterin22].
RESULTS
Epidemiological description of S. dysenteriae isolates
S. dysenteriae isolates were recovered from paediatric cohort, paediatric clinic- and hospital-based surveillance studies (Table 1). In total, 40 children with diarrhoea were culture-positive for Shigella. Twenty-one of the children were enrolled in the paediatric cohort studies and 19 were enrolled in clinic- and hospital-based studies for paediatric severe diarrhoea. The median age of children with S. dysenteriae was 15 [interquartile range (IQR) 9–23] months; 55% (n=22) were males and all infections occurred in the warm months of May to October.
Table 1. Identification and clinical characteristics of paediatric patients with Shigella dysenteriae (n=40) or Shigella boydii (n=30) isolated among predominate serotypes
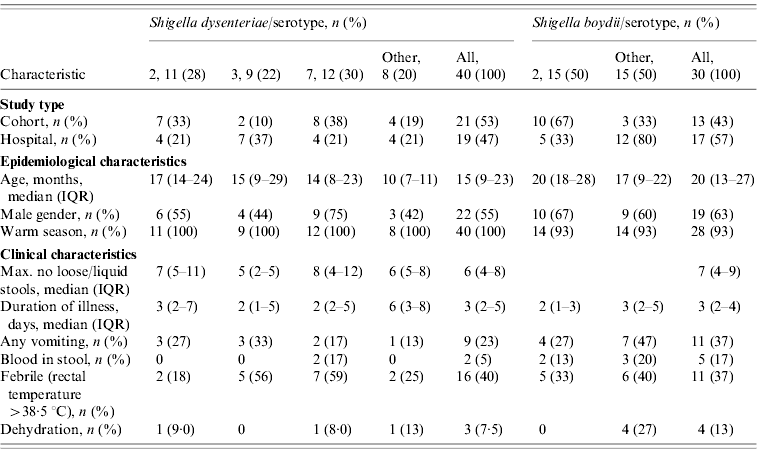
The S. dysenteriae-associated children presented with fever (n=16, 40%), vomiting (n=9, 22·5%), dehydration (n=3, 7·5%), and visible blood in stool (n=2, 5%). Three cases reported the use of antibiotics before the stool sample was taken. The median maximum number of loose stool during any diarrhoeal day was 6 (IQR 4–8), while the median duration of a diarrhoeal episode was 3 (IQR 2–5) days. ETEC and rotavirus were isolated as co-pathogens in six and two cases, respectively.
Overall, 43 S. dysenteriae strains were isolated from diarrhoeal stool samples from 40 children. Three children had pairs of S. dysenteriae isolates, recovered during the same diarrhoeal episode; however, only the first isolate was included in the clinical descriptive and genetic analyses (Table 1).
Epidemiological description of S. boydii cases
Similar to S. dysenteriae, isolates of S. boydii were recovered from all surveillance study sites during the warm season (93%, n=28) (Table 1).
A total of 31 isolates of S. boydii were recovered from the 30 diarrhoeal children. Two S. boydii isolates were isolated from one child from the same diarrhoeal episode; only the first isolate was analysed. Of the 30 children, 13 (43%) were from the paediatric cohort and 17 (57%) from the paediatric severe diarrhoea study.
The median age for all children with S. boydii infection was 20 (IQR 13–27) months and 63% (n=19) were males. ETEC was isolated as a co-pathogen in 8/30 (27%) samples while rotavirus and Campylobacter infection were co-pathogens in 14% (3/24) and 3% (1/30) of tested samples, respectively.
Clinical characteristics of diarrhoeal episodes due to S. boydii included vomiting (n=11, 37%), fever (n=11, 37%), and visible blood in stool (n=5, 17%). Dehydration was detected in 13% (n=4) of episodes. Antibiotics were used during 7% (n=2) of the episodes. The median duration of diarrhoeal episodes was 3 (IQR 2–4) days and the median maximum number of loose stools passed was 7 (IQR 5–9) times per day. Children with S. dysenteriae and/or S. boydii diarrhoea were more likely to have vomiting when presenting to hospital for care (P=0·005).
Diversity of S. dysenteriae and S. boydii isolate serotypes
Of 40 children infected with S. dysenteriae isolates, three serotypes [serotypes 7 (30%), 2 (27·5%), and 3 (22·5%)] accounted for the majority (80%) of all isolates (Table 1). Single isolates of S. dysenteriae serotypes 1 and 12 were identified during the 7 years of sample collection. Seventy-five percent of children infected with S. dysenteriae serotype 7 were male and their most common symptoms were fever (59%), vomiting (17%) blood in stool (17%) and dehydration (8%). However, 44% of children infected with S. dysenteriae serotype 3 were males and presented with fever (56%) and vomiting (33%). All seven S. dysenteriae serotypes found in this study were recovered from hospital and clinic study samples while only four serotypes and the untypable isolates (serotypes 2, 3, 4, 7) were present in the cohort studies.
Of the 30 S. boydii isolates, nine serotypes were identified; serotype 2 was the most common serotype (n=15, 50%) and was isolated in 67% of males. Children infected with S. boydii serotype 2 presented with fever (33%), vomiting (27%), blood in stool (13%), although none of the patients were reported as dehydrated. All nine S. boydii serotypes recovered were found in hospital samples while only three serotypes (serotypes 2, 4, 14) were found in the cohort studies. However, there was no significant difference in the distribution of serotypes among children with S. dysenteriae and S. boydii enrolled from hospital or from cohort studies.
Antimicrobial resistance profiles of S. dysenteriae and S. boydii
S. dysenteriae isolates showed high levels of resistance against TE (58%), AM (52%), C (42%), and SXT (25%, Table 2). Resistance against CR and NA was detected in only 12% and 5% of the isolates, respectively. Only 12 isolates were susceptible to all antibiotics tested, and eight isolates were resistant to either one (n=4) or two (n=4) antibiotics. The majority of S. dysenteriae serotype 2 isolates were susceptible (9/11) to all agents tested. From the 40 S. dysenteriae isolates, half (n=20, 50%) were MDR. The majority of MDR were resistant to three antibiotics (n=16), although three isolates were resistant to four and a single isolate was resistant to six antibiotics. All serotype 3 isolates (n=9) were MDR; all were resistant to AM, C and TE. Isolates expressing serotype 7 showed more antimicrobial diversity; three isolates were completely susceptible, a single isolate was resistant to two antibiotics, and eight isolates were MDR. No ESBL-producing isolates were detected in either the S. dysenteriae or S. boydii collection.
Table 2. Antimicrobial resistance results for S. dysenteriae and S. boydii isolates from Egypt, 1999–2006
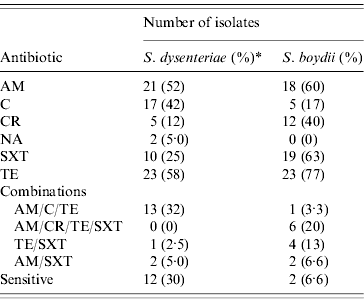
AM, Ampicillin; C, chloramphenicol; CR, cephalothin; NA, nalidixic acid; SXT, sulfatrimethoxazole; TE, tetracycline.
* The percentage of resistant isolates is shown within parentheses; for S. dysenteriae this percentage was calculated with a denominator of 40 isolates; for S. boydii, this percentage was calculated with a denominator of 30 isolates.
Isolates of S. boydii were highly resistant to TE (77%), SXT (63%), AM (60%) and CR (40%); while less resistant to C (17%), and all strains were susceptible to NA (Table 2). The most common MDR phenotypic profile (40%, n=12) detected in S. boydii were isolates resistant to the combination of AM, CR, TE, and SXT (20%). Over half of the S. boydii isolates were MDR (n=16, 53%); nine isolates were resistant to a combination of four antibiotics, and six isolates were resistant to three agents. Only one isolate was resistant to five antibiotics. Similarly, only two isolates were sensitive to all the antimicrobials tested in this study. We found six isolates resistant to either one or two antimicrobial agents. Neither S. boydii nor S. dysenteriae showed resistance to CIP.
Genetic basis of AM and SXT resistance
To explore whether there were genetic changes associated in the resistance mechanisms over time, we screened all Shigella isolates for genes involved in resistance to AM and SXT (Table 3). Evidence for redundant AM resistance (AM-R) in S. dysenteriae was apparent; 18/21 (86%) isolates carried a bla OXA-7-type and 17/21 (81%) harboured a bla TEM-1-type β-lactamase. In contrast, 8/18 (44%) of AM-R S. boydii isolates carried bla OXA-7-type and all (n=18) harboured a bla TEM-1-type β-lactamase. Only 10 (25%) S. dysenteriae isolates were resistant to SXT and the majority of those isolates harboured the sulI (n=9) and/or sulII (n=8) genes. Moreover, the majority of the SXT-R S. boydii isolates carried sulI (n=14, 74%) and/or sulII (n=17, 89%).
Table 3. Carriage of antimicrobial resistance genes to β-lactams and sulfonamides among Shigella-resistant isolates
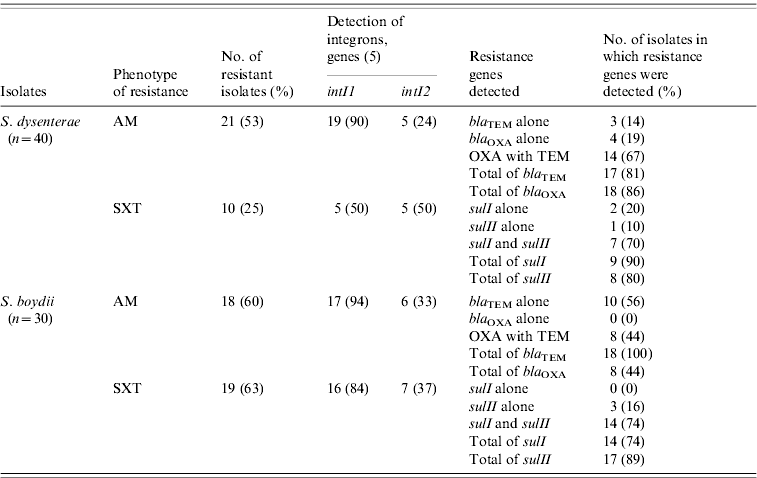
AM, ampicillin; SXT, sulfatrimethoxazole.
In AM-R S. dysenteriae, class I integrons (intI1) were predominant (n=19, 90%); while class II integrons (intI2) were detected in only 24% (n=5) of these isolates (Table 3). However, the genes intI1 or intI2 were detected in only 50% of the S. dysenteriae SXT-R isolates. Most of the resistant S. boydii carried a marker for intI1. For example, intI1 was detected in the majority of resistant isolates for AM (n=17, 94%) and SXT (n=16, 84%). In contrast, intI2 was detected only in 33% (n=6) AM-R and 37% SXT-R (n=7) isolates.
XbaI–PFGE confirms species diversity
XbaI–PFGE of S. dysenteriae isolates generated 28 unique profiles (Fig. 1). We selected a similarity value of 47% because at this similarity value, isolates separated into the three clusters mainly had the same serotype. At this cut-off value, all of the S. dysenteriae serotype 2 isolates (n=11) clustered together (cluster 1). Two untypable isolates, the sole serotype 12 isolate, and single serotype 3 isolate were also in this cluster. Cluster 2 consisted of all the serotype 7 (n=12) and a single serotype 1 isolates. Cluster 3 consisted of all serotype 4 and all but one of the serotype 3 isolates (n=8). Considering a similarity value of 64%, seven subclusters of isolates were evident. Three subclusters fell within cluster 1; one subcluster was strictly populated by serotype 2 isolates; one was composed of only the untypable isolates and serotype 2; the last subcluster was mixed with serotype 2, 3 and 12 isolates. Cluster 2 was composed of two subclusters, the first subcluster strictly populated by serotype 7 (n=11); the second subcluster was composed of single isolates expressing either serotype 1 or serotype 7. Cluster 3 contained two subclusters, one subcluster composed of serotype 3 exclusively, a second subcluster contained a single serotype 3 isolate and an indistinguishable set of serotype 4 isolates.

Fig. 1. Dendrogram showing the relationship of S. dysenteriae isolates based on XbaI–PFGE profiles. Similarity of banding patterns of XbaI-restricted whole genomic DNA from isolates of S. dysenteriae were compared using Bionumerics software. The bar at the top of the figure indicates decreasing genetic similarity (moving from the right hand to the left hand side of the figure). Numbers located at nodes within the dendrogram indicate percent similarity. Isolates designated as ‘WS’ were recovered from the paediatric birth cohort; isolates designated as ‘HS’, ‘MH’, or ‘PS’ were recovered from children seeking medical care for diarrhoea-related symptoms. Clusters of isolates determined at a 47% band pattern similarity are shown in the boxes. Other abbreviations: GR, serogroup; TY, serotype; AST, antibiotic sensitivity testing result (indicating resistance to a specific antibiotic); Date-R, specimen receipt date; Ne, did not react with antisera; AM, ampicillin; C, chloramphenicol; CR, cephalothin; NA, nalidixic acid; SXT, sulfatrimethoxazole; TE, tetracycline.
The number of S. dysenteriae isolates with MDR profiles was significantly different based on the different studies (Fig. 1). For example, 65% of isolates from the hospital studies were MDR compared to 35% from the cohort studies (P=0·028); however, MDR status was not associated with any specific sign or symptom. MDR status was associated with serotype distribution, (P=0·0001); 100% of serotype 3 and 67% of serotype 7 were MDR.
Characterization of S. boydii using XbaI–PFGE (Fig. 2) identified 25 profiles. The S. boydii population had a different structure to that observed with S. dysenteriae and at a 54% similarity level, six clusters of isolates, mainly in agreement with the serotyping results, were evident. Cluster 1 and cluster 2 were single isolates of serotype 5 or serotype 13, respectively. Cluster 3 was the largest cluster, containing all of the serotype 2 isolates (n=15), the single serotype 1 isolate and one serotype 4 isolate. Cluster 4 contained both serotype 18 isolates, and one of the two serotype 4 isolates. Cluster 5 consisted of all four untypable isolates and the sole serotype 17 isolate. The final cluster (cluster 6) consisted of all the serotype 14 isolates. Increasing the percentage of similarity to 70% had the greatest impact on isolates in cluster 3. At this similarity level, three subclusters were identified in cluster 3; one subcluster consisted of the serotype 1 and serotype 4 isolates; a second closely related subcluster consisted of a set of four genetically indistinguishable serotype 2 isolates that spanned multiple years; the third subcluster was composed only of highly related serotype 2 isolates (n=11) that also spanned multiple years. Cluster 4 contained serotype 18 isolates and the serotype 4 isolate as one group, cluster 5 split into two subgroups; the first group consisted of two untypable strains, and the sole serotype 17 isolate; the second subgroup consisted of the remaining two untypable isolates. Cluster 6 also split into two subclusters, representing potential independent lineages of serotype 14.
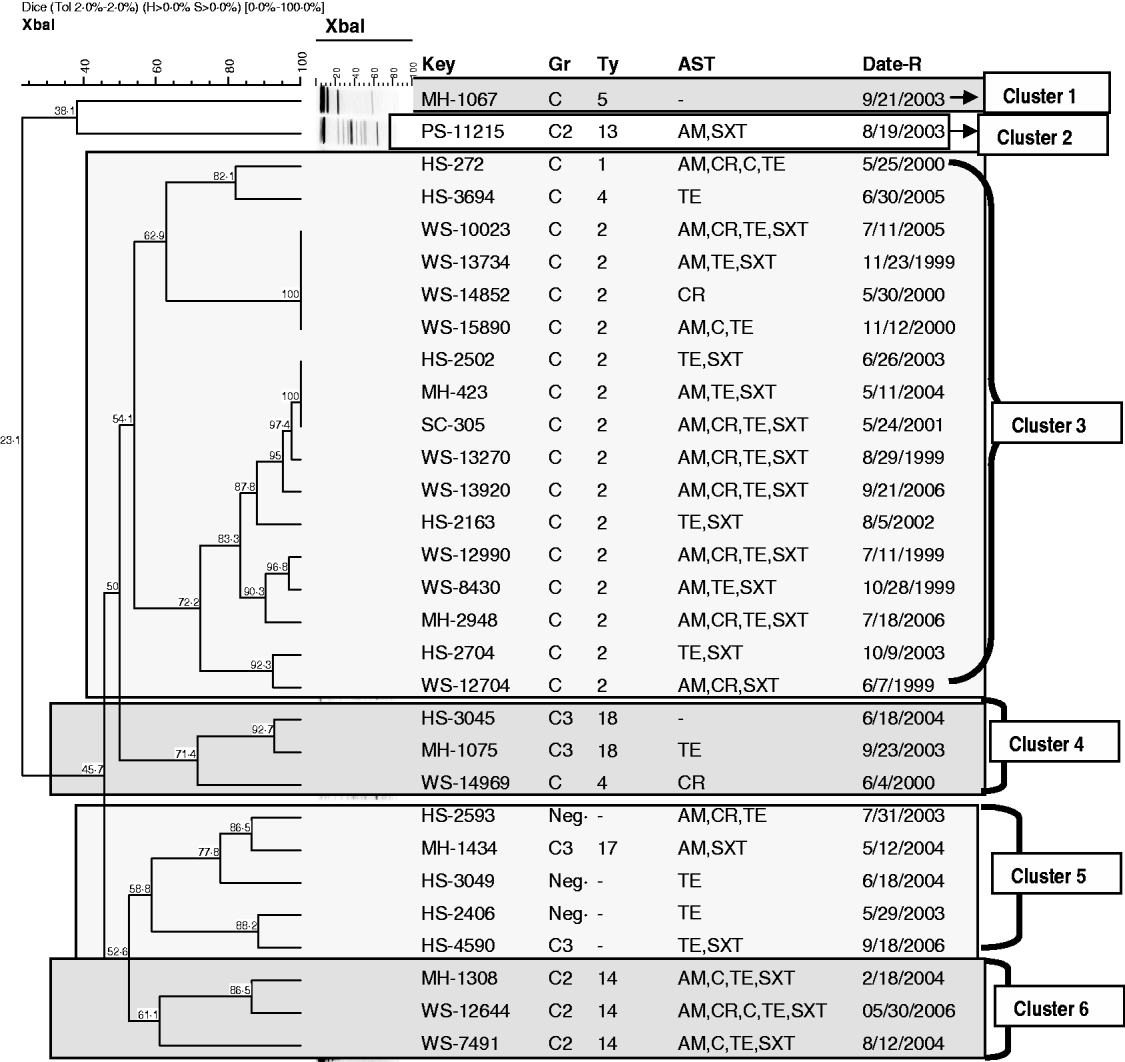
Fig. 2. Dendrogram showing the relationship of S. boydii isolates based on XbaI–PFGE profiles. Similarity of banding patterns of XbaI-restricted whole genomic DNA from isolates of S. dysenteriae were compared using Bionumerics software. The bar at the top of the figure indicates decreasing genetic similarity (moving from the right hand to the left hand side of the figure). Numbers located at nodes within the dendrogram indicate percent similarity. Isolates designated as ‘WS’ were recovered from the paediatric birth cohort; isolates designated as ‘HS’, ‘MH’, or ‘PS’ were recovered from children seeking medical care for diarrhoea-related symptoms. Clusters of isolates determined at a 54% band pattern similarity are shown in the boxes. Other abbreviations: GR, serogroup; TY, serotype; AST, antibiotic sensitivity testing result (indicating resistance to a specific antibiotic); Date-R, specimen receipt date; Ne, did not react with antisera; AM, ampicillin; C, chloramphenicol; CR, cephalothin; NA, nalidixic acid; SXT, sulfatrimethoxazole; TE, tetracycline.
S. boydii MDR isolates were found more frequently in the paediatric cohort samples (Fig. 2). A total of 10/12 (83%) S. boydii isolates from the paediatric cohort were MDR; only 6/18 (33%) hospital samples were MDR. S. boydii serotype 2 (n=15) were often associated with MDR (11/15, 73%).
DISCUSSION
Despite the potential significance of explosive outbreaks of dysentery [Reference Kernéis23] and the spread of drug-resistant strains through the consumption or use of contaminated food [Reference Löfdahl24] or water sources [Reference Smith25], there is a paucity of data from Egypt regarding the characterization of non-flexneri Shigella. S. dysenteriae and S. boydii are often the least common serogroups causing shigellosis in developing and developed countries and neither are considered common causes of diarrhoea. Nevertheless, outbreaks of both species have been reported. In a recent report from Sweden, an outbreak of S. dysenteriae serotype 2 was linked to sugar snap peas, most probably originating from Kenya, and passing through at least one other European country [Reference Löfdahl24]. Similarly, a cluster of cases of S. boydii serotype 2 linked to water transmission was recently reported in Mpumalanga Province in South Africa [Reference Smith25].
In our first report [Reference Ahmed11] we presented data from children infected with S. flexneri. In this report we present data from children infected by S. dysenteriae and S. boydii exclusively. Data with respect to diarrhoea associated with S. sonnei will be reported separately. In this report, we have characterized collections of S. dysenteriae and S. boydii isolates recovered from children aged <5 years living in Egypt who were enrolled in one of three paediatric birth cohort studies or a paediatric severe diarrhoea study from 1999 to 2006. Wierzba et al. [Reference Wierzba10] reported previously that the dominant bacterial pathogen present in the studies from which our Shigella isolates were recovered was ETEC, distantly followed by Campylobacter spp. The third most common bacterial pathogen was Shigella spp. and S. flexneri was the most abundant species isolated from these studies. A study of severe diarrhoeal cases in Dhaka, Bangladesh [Reference Khan26] conducted over 18 months from 2000 to 2001 (all-age diarrhoea) indicated a similar distribution of S. dysenteriae (n=20) and S. boydii (n=16) isolates.
As a common concept among physicians in rural clinics, shigellosis is suspected and treated when fever and bloody stool are associated with diarrhoea; however, in our study only 5% and 17% of children with S. dysenteriae- and S. boydii-associated diarrhoea, respectively, had bloody stools; this finding is consistent with previous cohorts and hospital-based studies from Egypt [Reference Wasfy8]. Children enrolled in the cohort studies had mainly mild to moderate diarrhoea episodes but those presenting to a medical care facility were more severe as indicated by a significant percentage of vomiting and dehydration. These findings indicate that the clinical presentation of shigellosis due to either S. boydii or S. dysenteriae can range from mild to severe illness. We also noted greater diversity in isolates of both pathogens presenting to a medical care provider, as determined by serology and XbaI–PFGE typing; one possible explanation of this observation is that children presenting to a medical provider were from different geographical areas throughout Egypt while children enrolled in the cohort studies were restricted to a cluster of villages in one governorate in Egypt. A separate observation for which we do not yet have an adequate explanation relates to antibiotic resistance. Perhaps, as expected, the majority of S. dysenteriae MDR isolates were present in children presenting to medical providers. This may reflect prior antibiotic use due to the severity of observed disease compared to children in the cohort study. However, the observation of a greater number of S. boydii MDR cases in the paediatric cohort, rather than presenting to a medical care provider, is puzzling. Further study is necessary to understand this observation.
Immunity against Shigella is serotype specific and there is limited cross-protection between the various serotypes [Reference Kotloff2]. S. dysenteriae isolates were mainly divided into expression of three serotypes (7, 2, 3). Only a single S. dysenteriae serotype 1 was isolated. In contrast, we found that the majority of S. boydii isolates expressed serotype 2 (50%). The next most prevalent serotype was 14 (10%). This would suggest the use of a multivalent vaccine could effectively control Shigella spp. in Egypt.
We also assessed the serotype distribution of S. dysenteriae and S. boydii from Egypt to determine whether it was unique from that of other countries with endemic shigellosis. Nearly all of the S. dysenteriae isolates were able to be serotyped using commercial antisera (95%); relatively speaking, a much higher percentage of S. boydii isolates were untypable (13%). A recent study from Karachi also examined the distribution of Shigella serotypes in an endemic setting [Reference Zafar27]. This group reported a higher percentage for untypable S. dysenteriae isolates (16%) although they had a similar percentage for untypable S. boydii isolates (12%) comparing a similar number of S. dysenteriae and S. boydii isolates to our study. S. dysenteriae serotype 7-expressing isolates were most common in Pakistan (24%) in partial agreement with our results (30%). However, our results showed that the distribution of serotypes for both species was different. In Pakistan, S. dysenteriae serotype 7-expressing isolates were most common, followed by serotypes 4, 2 and 12. Three S. boydii serotypes, 1, 2, and 8, were equally abundant (each 16%) in Karachi.
We observed that AM resistance among S. dysenteriae (52%) was slightly lower than S. boydii (60%). Different results for AM-resistant isolates were found among S. dysenteriae (37%) and S. boydii (25%) in six Asian countries in 2006 [Reference Von Seidlin28], while higher resistances were reported in Pakistan in 2009, for S. dysenteriae (68%) and S. boydii (35%) [Reference Zafar27], respectively. In this study, susceptibility to NA in S. dysenteriae and S. boydii was high (95% and 100%, respectively) and all our isolates were susceptible to CIP. This agrees with reports from Yemen, Iran and Turkey [Reference Al-Moyed29–Reference Farshad31]. In contrast, the resistance profile in India and Bangladesh is worrisome. NA resistance among S. dysenteriae isolates in at least one study in India was alarming (82%), and over half (54·5%) of these isolates were CIP resistant [Reference Taneja32]. While not isolated at as great a frequency, S. boydii isolates in this study also demonstrated an elevated resistance to NA (57·1%) although CIP resistance was not detected (0%). Although Bangladesh has reported an overall decrease in isolation of Shigella over the last 15 years, there has been a rise in the resistance to all WHO-recommended first-line antimicrobial therapies for shigellosis during this time [Reference Khatun33]. In Egypt, quinolones are not generally recommended for use in children and this may contribute to the low levels of observed resistance. Other possible factors contributing to the infrequent observation of fluoroquinolone resistance in Egypt may be the relatively high cost and lack of availability. A study measuring the risk factors associated with quinolone resistance in the region could contribute to our understanding of the observed differences.
Occurrence of MDR Shigella spp. strains is of great public health concern. In Egypt, Wasfy and colleagues [Reference Wasfy8] looked at sensitivities to 12 antibiotics; their findings with respect to S. dysenteriae indicated that from the 33 isolates analysed, 86% were resistant to erythromycin, 68% to C, 56% to TE, 55% to AM, while only 11% of isolates were resistant to SXT. No isolates were resistant to NA and CIP resistance was not tested. Analysing data from the paediatric cohorts, Putnam et al. [Reference Putnam6] observed that S. dysenteriae isolates cultured from 1995 to 2000 showed little resistance to AM, C, SXT, or TE and only 2/28 isolates were MDR. Our results appear to be more similar to those of Wasfy and colleagues [Reference Wasfy8]; 48% of our S. dysenteriae isolates were MDR (19/40) and 43% of these isolates were resistant to AM, C, and TE. Resistance levels to AM (53%) and TE (58%) remained at nearly 50%, but we observed less resistance to C (43%). In a separate analysis, Wasfy et al. [Reference Wasfy8] suggested that a decrease in the use of C for the treatment of typhoid fever has led to an increased susceptibility of the causative organism; it is possible that a similar change in antimicrobial therapy is having the same effect for isolates of S. dysenteriae.
Our findings indicate that the resistance genes of sulfonamides (sulI and sulII) and β-lactams (bla TEM, and bla OXA) are widely disseminated in S. dysenteriae and S. boydii isolates in Egypt and many of these isolates carry integrons, potentially providing them with the capability of obtaining antibiotic resistance. Little information on the integron content of S. boydii and S. dysenteriae isolates is available, but in our study, harbouring a class 1 integron among either Shigella spp. was associated with AM resistance, in agreement with results reported previously [Reference Dubois34]. Class II integrons (intI2) were detected in only 24% of S. dysenteriae and 33% of S. boydii AM-resistant isolates. In addition, intI1 or intI2 was detected almost equally in S. dysenteriae SXT-resistant isolates. Among S. boydii SXT-resistant isolates, intI1 detection (84%) was higher than intI2 (37%). The high prevalence of the OXA-1-type and TEM-1-type β-lactamases in S. dysenteriae have been reported previously [Reference Dubois34] and is also in agreement with our results (86% and 81%, respectively).
PFGE has been widely used for typing Shigella spp. and is considered a molecular typing tool with high discriminatory power for detection of outbreaks [Reference Löfdahl24, Reference Gerner-Smidt35] and longitudinal observation of bacterial populations [Reference Sánchez36, Reference Talukder37]. We observed a variety of pulsed-field types present in both S. dysenteriae and S. boydii. Comparative analysis of PFGE pulsotypes indicated that most observed clusters also corresponded to the expression of a specific lipopolysaccharide antigen, as determined by their serotyping reaction. This observation was also true in our previous study investigating the diversity in S. flexneri isolates [Reference Ahmed11]. PFGE results in this study showed six sets of S. dysenteriae isolates (n=18) and two sets of S. boydii isolates (n=7) that were indistinguishable and with the exception of one S. dysenteriae group, the isolates were not epidemiologically linked. These data would appear to indicate that some isolates of S. dysenteriae and S. boydii are capable of persisting in the environment over time. An intriguing hypothesis that remains to be tested is whether Shigella isolates in Egypt might persist in the environment by colonizing Acanthamoeba spp., similar to a mechanism proposed for S. sonnei [Reference Jeong38]. Acanthamoeba spp., including the pathogenic T4 genotype, have been documented in the Nile Delta region, and were found associated in areas where cooking, cleaning, washing and fishing took place [Reference Lorenzo-Morales39].
In this study, we have shown that there is rich serological and genetic diversity among isolates of S. dysenteriae and S. boydii. Some limitations of the study are the low numbers of isolated S. dysenteriae and S. boydii which made determination of statistical significance difficult and our inability to perform follow-up investigations on children seeking medical care in Abu Homos, Fayoum and Moqqatum Hills. Any ongoing vaccine development against shigellosis should take into consideration the amount of variability across multiple geographical locations including Egypt. At least one group of investigators are in advanced clinical trials of a live oral Shigella vaccine; it is important to support these trials with additional diarrhoea surveillance studies in other governorates within Egypt and other high-risk countries, to monitor serotype distribution and to determine potential vaccine coverage. In addition, we have observed antibiotic resistance levels and mechanisms of resistance which can be used as a basis for comparison in future studies in Egypt and the Middle East. This study provides useful information for future surveillance of shigellosis and implementation of antibiotic treatment policies in Egypt and elsewhere in the region.
ACKNOWLEDGEMENTS
The opinions or assertions herein are the private views of the authors and should not be construed as official or as reflecting the views of the United States Department of the Navy, Department of Defense, nor the United States Government. This study [DoD no. NAMRU3.2000.0002 (IRB Protocol No. 96), NAMRU3.1997.0002 (IRB No. 72), NAMRU3.1998.0003 (IRB No. 75), NAMRU3.2003.0011 (IRB No. 145)] was reviewed and approved by the Institutional Review Boards of the US Naval Medical Research Unit No. 3 and the Egyptian Ministry of Health in compliance with all Federal regulations governing the protection of human subjects. Informed consents were obtained from all adult participants and from parents or legal guardians of minors.
DECLARATION OF INTEREST
None.







