Introduction
Dunaliella (Chlorophyceae, Dunaliellales) is a genus of algae with immense economic potential owing to its production of an array of exploitable compounds, including β-carotene, glycerol and phytosterols (reviewed by Avron & Ben-Amotz, Reference Avron and Ben-Amotz1992). Species within this genus include halophilic and halotolerant strains and are frequently encountered in hypersaline environments. Dunaliella lacks a rigid cell wall and has a flexible cell membrane capable of rapidly changing shape in response to osmotic stress (Oliveira et al., Reference Oliveira, Bisalputra and Antia1980) and this can confound morphological identification. Cell size is highly variable with differences being related to growth conditions, e.g. nutrients, light intensity and salt concentration (Borowitzka & Brown, Reference Borowitzka and Brown1974), and can also vary within the same culture, for example D. salina (Teodoresco, Reference Teodoresco1905) can exhibit 5–29 μm cell length and 3.8–20.3 μm width (Borowitzka & Siva, Reference Borowitzka and Siva2007).
Early observations and descriptions of the genus Dunaliella laid the foundations of a well characterised group (Teodoresco, Reference Teodoresco1905; Lerche, Reference Lerche1937; Massjuk, Reference Massjuk1973) with the family Dunaliellaceae comprising of four sections of Dunaliella, as described by Massjuk (Reference Massjuk1973); section Tertiolectae which are oligo-euhaline, do not accumulate carotenes and grow at an optimum salinity of <6% NaCl; section Dunaliella which are halophilic species that accumulate carotenes; section Virides which are hyperhaline, always green and radially symmetrical; and Peirceinae which are hyperhaline, always green but cells are bilaterally symmetrical. Presently, within the section Dunaliella are three accepted species, D. salina, D. parva and D. pseudosalina as well as D. bardawil. The latter has been debated as to whether it should actually be deemed to be a separate species, or is in fact D. salina considering its characteristics match the species description originally specified by Teodoresco (Reference Teodoresco1905) (Borowitzka & Siva, Reference Borowitzka and Siva2007; González et al., Reference González, Gómez, Polle, Ben-Amotz, Polle and Subba Rao2009).
Dunaliella salina has been found to be able to tolerate salt saturation of approximately 5.5 M NaCl (reviewed by Avron & Ben-Amotz, Reference Avron and Ben-Amotz1992). It is a particularly polymorphic species, with cell physiology related to abiotic factors. Cells can be pigmented green or red depending on the amount of β-carotene accumulated and under high stress conditions, members of this species can accumulate >5% β-carotene dry weight (Ben-Amotz & Avron, Reference Ben-Amotz and Avron1983). D. bardawil was isolated from a salt pond near Bardawil, Israel in 1976 and was reported to accumulate considerably larger amounts of β-carotene compared to D. salina, accumulating β-carotene in membrane-free globules in the interthylakoid spaces of the chloroplast (Ben-Amotz et al., Reference Ben-Amotz, Katz and Avron1982). However, it is now unclear if the strain of D. salina used to compare to D. bardawil in this study was a good representative of D. salina, as subsequent reports have indicated that D. bardawil UTEX (Culture Collection of Algae at the University of Texas at Austin) 2538 and D. salina UTEX 1644 actually have a similar carotenoid: chlorophyll ratio (Jahnke, Reference Jahnke1999). Morphologically it has proven difficult to resolve differences between strains of D. salina, however, four formae have previously been described in the literature. D. salina spp. salina fo. sibirica; a species with cells that are broader in the median or anterior region, D. salina spp. salina fo. oblonga; cylindrical cells of 7–28 μm length, 5–13 μm width, D. salina spp. salina fo. magna; cells ovoid with a cell volume >1000 μm3 and cell length 7.5–29 μm, cell width 7.5–21 μm, and D. salina spp. salina fo. salina; which is similar to magna but with smaller cell dimensions (Massjuk & Radchênko, Reference Massjuk and Radchenko1973). These formae have primarily been based on cell shape which can be an ambiguous criterion to use as cell shape can vary considerably based on differing culture conditions.
Genotyping is now considered imperative in aiding the classification of Dunaliella species, with the ribosomal markers at the forefront of phylogenetic analyses (e.g. González et al., Reference González, Coleman, Gómez and Montoya2001; Olmos et al., Reference Olmos, Ochoa, Paniagua-Michel and Contreras2009; Bucheim et al., Reference Bucheim, Kirkwood, Bucheim, Verghese and Henley2010; Assunção et al., Reference Assunção, Jaén-Molina, Caujapé-Castells, de la Jara, Carmona, Freijanes and Mendoza2012), however, other commonly used markers for Dunaliella sp. and, indeed other marine algae, include rbcL (Preetha et al., Reference Preetha, John, Subin and Vijayan2012) and tufA (reviewed by Leliaert et al., Reference Leliaert, Verbruggen, Vanormelingen, Steen, Lopez-Bautista, Zuccarello and De Clerck2014). Furthermore, by exploiting the sporadic occurrence of the group I introns within the 18S nuclear SSU rDNA (Wilcox et al., Reference Wilcox, Lewis, Fuerst and Floyd1992), the SSU marker has been used as a size indicator of different Dunaliella species (Olmos-Soto et al., Reference Olmos-Soto, Paniagua-Michel, Contreras and Trujillo2002). The absence of introns in the SSU rDNA in the Tertiolectae, one intron in D. salina, two introns in D. bardawil and one intron for D. viridis (that differs to the aforementioned species), have been used as an aid in identifying these species. Moreover, Olmos et al. (Reference Olmos, Ochoa, Paniagua-Michel and Contreras2009) also used the method as a way to distinguish hyper-producers of β-carotene from other Dunaliella sp. and to discriminate between D. salina var Teod and D. salina/bardawil.
The hypervariable regions, ITS 1, 5.8S rRNA and ITS 2, have been frequently employed by molecular studies of this important algal group (e.g. González et al., Reference González, Coleman, Gómez and Montoya2001) with some studies focussing on the ITS 2 spacer sequence (Assunção et al., Reference Assunção, Jaén-Molina, Caujapé-Castells, de la Jara, Carmona, Freijanes and Mendoza2012). Assunção et al. (Reference Assunção, Jaén-Molina, Caujapé-Castells, de la Jara, Carmona, Freijanes and Mendoza2012) undertook a comprehensive analysis of Dunaliella species using ITS 2, and identified three main clades within the section Dunaliella; salina I, salina II, and pseudosalina. Following on from this, however, Assunção et al. (Reference Assunção, Jaén-Molina, Caujapé-Castells, Wolf, Bucheim, de la Jara, Freijanes, Carmona and Mendoza2013) allied the pseudosalina clade as putative D. viridis and furthermore, in agreement, Borowitzka & Siva (Reference Borowitzka and Siva2007), had also previously re-classified members of the pseudosalina clade based on morphology. Notably, they reassigned D. salina CCAP 19/3 as D. viridis, D. parva SAG 19-1 as D. maritima and D. salina UTEX 200 as D. viridis making the pseudosalina clade indeterminate. Assunção et al. (Reference Assunção, Jaén-Molina, Caujapé-Castells, Wolf, Bucheim, de la Jara, Freijanes, Carmona and Mendoza2013) concluded that many of the taxonomic assignments of Dunaliella strains based on morphological and physiological measurements were flawed and sequence analysis of the ITS 2 was required to affirm the taxonomic affiliation of numerous strains. Moreover, it appeared that some strains sequenced were different to what they were originally designated, potentially as a result of cross-contamination. ITS markers are often favoured as there are many copies in the genome making it easy to amplify and insertions/deletions within the sequence are common meaning there is good variability between species. Other nuclear markers used for the analysis of Dunaliella from hypersaline environments include the large subunit rDNA and small subunit rDNA (Bucheim et al., Reference Bucheim, Kirkwood, Bucheim, Verghese and Henley2010) and these have shown good potential for taxonomic resolution.
In addition to sequencing and phylogeny of the ITS 2, analysis of compensatory base changes (CBC) in the rRNA secondary structure of this region has also been shown to be useful in identifying organisms that are distinct species. A minimum of one CBC (with 93% confidence in plants and fungi) between two organisms can designate them as separate species (Muller et al., Reference Müller, Philippi, Dandekar, Schultz and Wolf2007). Notably, however, the absence of CBCs does not necessarily mean that they are the same species. Assunção et al. (Reference Assunção, Jaén-Molina, Caujapé-Castells, de la Jara, Carmona, Freijanes and Mendoza2012) used this method in their investigations of Dunaliella sp. but found its application to the delineation of the group limited due to a lack of CBCs identified in isolates known to be distinct species.
Both rbcL and tufA are plastid genes, with tufA encoding elongation factor Tu, and has become more frequently used for molecular studies of algae (Cook et al., Reference Cook, Whittock, Wright and Hallegraeff2011; Moniz et al., Reference Moniz, Guiry and Rindi2014). Availability of sequences, however, is limited making phylogenetic analysis less conclusive compared to other markers. Moniz et al. (Reference Moniz, Guiry and Rindi2014) used the tufA gene in their analysis of the order Prasiolales (Chlorophyta) finding that there was good agreement between the phylogenies of tufA, rbcL and psaB. Presently, tufA as a marker for Dunaliella taxonomy has not been thoroughly examined and its potential not fully realised. rbcL analysis has typically not highlighted intraspecific variation to the same degree as other markers such as the ITS regions. Preetha et al. (Reference Preetha, John, Subin and Vijayan2012) reported that rbcL gene phylogeny of Dunaliella did not show the same level of heterogeneity as the ITS, yet, the novel isolates analysed in their study showed similar groupings in both phylogenetic trees.
In addition to these useful barcoding markers for clonal isolates which can also be used for environmental analysis, the V9 variable region of the SSU rRNA, has frequently been employed for next generation sequencing of environmental samples (Amaral-Zettler et al., Reference Amaral-Zettler, McCliment, Ducklow and Huse2009; de Vargas et al., Reference de Vargas, Audic, Henry, Decelle, Mahé, Logares, Lara, Berney, Le Bescot, Probert, Carmichael, Poulain, Romac, Colin, Aury, Bittner, Chaffron, Dunthorn, Engelen, Flegontova, Guidi, Horák, Jaillon, Lima-Mendez, Lukeš, Malviya, Morard, Mulot, Scalco, Siano, Vincent, Zingone, Dimier, Picheral, Searson, Kandels-Lewis, Acinas, Bork, Bowler, Gorsky, Grimsley, Hingamp, Iudicone, Not, Ogata, Pesant, Raes, Sieracki, Speich, Stemmann, Sunagawa, Weissenbach, Wincker and Karsenti2015) with an extensive database of sequences now available. The application of V9 to Dunaliella sp. taxonomy and its ability to resolve different species is therefore of importance to validate with regards to metagenomic diversity. To ensure this region can resolve the diversity of this group means that metagenomic analysis of hypersaline environments using this marker will accurately resolve the Dunaliella community diversity.
Suitable molecular markers are needed for accurate identification of species as this will aid in accurately identifying those isolates that will be economically valuable, e.g. strains of D. salina that produce high levels of 9-cis-β- carotene that is economically more valuable than its isomer all-trans-β-carotene, and to further understand the molecular evolution of this important group. This study set out to genetically investigate a range of new Dunaliella isolates collected from a range of geographical provinces including Israel, Spain, South Africa and Namibia. These were compared to reference strains previously collected from Chile, Australia, Mexico, Norway and Israel. We sought to employ a suite of molecular tools to provide a comprehensive analysis of different markers and their suitability for application to the genus Dunaliella.
Materials and methods
Dunaliella isolation and culture
Water samples (50 mL) were collected from hypersaline locations detailed in Table 1 and were transported back to the laboratory. Dunaliella sp. were identified via light microscopy and isolated using single cell-picking with a micropipette and dilution techniques according to Anderson & Kawachi (Reference Anderson, Kawachi and Anderson2005). Established cultures were maintained in 30 mL F/2 media (Guillard & Ryther, Reference Guillard and Ryther1962) with the addition of 50 g L−1 sea salts (Sigma) (1.45 M NaCl), at 25°C continuous light of 100 μmol m−2 s−1. Cultures were regularly sub-cultured into fresh media every 2–3 weeks and cell imaging was undertaken 2 weeks post sub-culture using a DMi8 live cell imaging system (Leica). All isolates have been deposited in the Marine Biological Association culture collection.
Table 1. Dunaliella sp. isolates used during this study
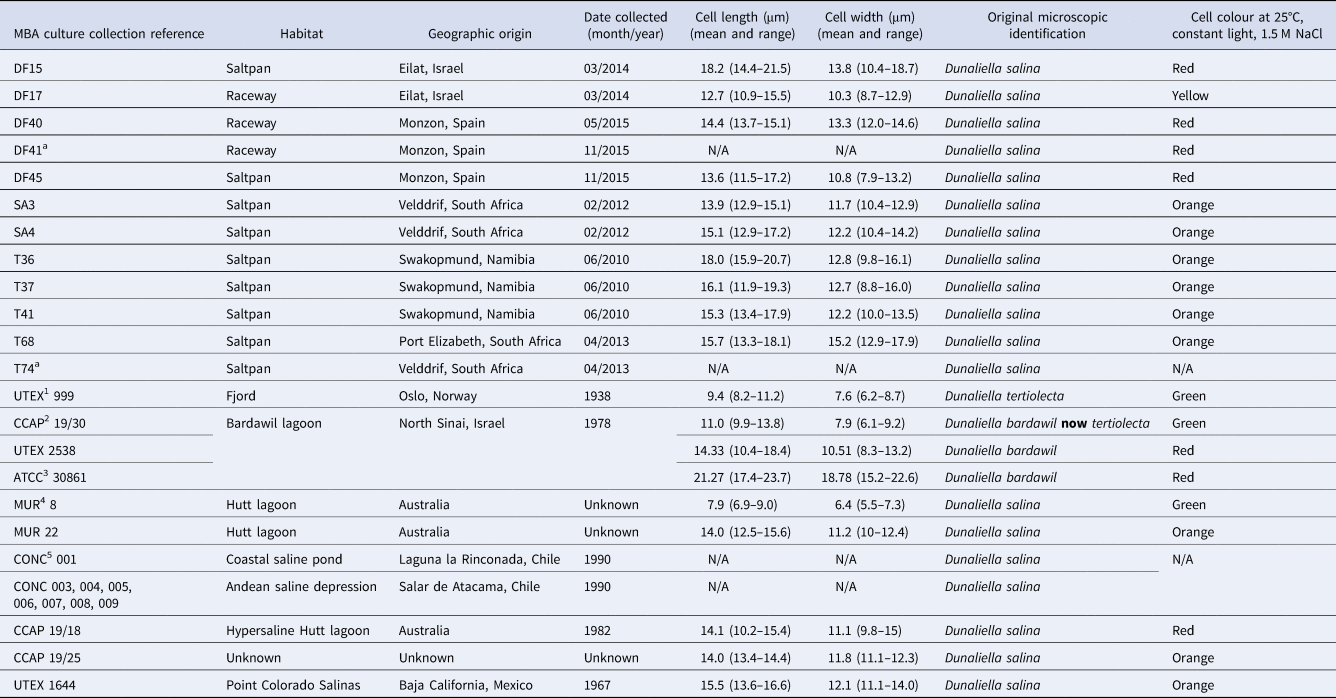
1 UTEX, Culture collection of Algae at the University of Texas, Austin, USA; 2CCAP, Culture collection of Algae and Protozoa, UK; 3ATCC, American type culture collection, VA, USA; 4MUR, Murdoch University Algal collection, Australia; 5CONC, Culture collection of Microalgae, Universidad de Concepcion, Concepcion, Chile.
a Isolates lost from culture.
DNA extraction, PCR and sequencing
DNA was extracted from 10 mL late exponential cultures using the DNeasy blood and tissue kit (Qiagen) according to the manufacturer's instructions with the exception of the elution volume which was 50 μL. PCR was carried out using a suite of primers (Table 2) in a Corbett Thermocycler (Corbett Research). PCR reactions were typically carried out in 50 μL volumes containing 2 μL DNA, 25 pmol each primer, 1 × reaction buffer, 2.5 mM MgCl2, 0.0025 mM dNTPs, 1 Unit Gotaq polymerase (Promega) unless otherwise stated (Table 2). PCR reactions proceeded with an initial denaturation at 95°C for 5 min, followed by 35 cycles of denaturation at 95°C for 30 s, annealing at 54°C for 45 s and extension at 72°C for 1 min unless otherwise stated (Table 2). PCR reactions had a final extension step of 72°C for 5 min. PCR products were either sequenced directly using the respective primers (Source Bioscience, Cambridge) or in some cases cloning was necessary to ensure a single sequence was obtained. In these instances, PCR products were electrophoresed on a 1.2% (w/v) agarose gel in 1 × TAE and purified using the Zymoclean gel purification kit (Cambridge Biosciences). Purified PCR product (1 μL) was ligated into the pCR2.1 TA cloning vector (Invitrogen) and transformed according to manufacturer's instructions. PCR amplicons from colony PCR with M13 primers were sequenced (Source Bioscience, Cambridge).
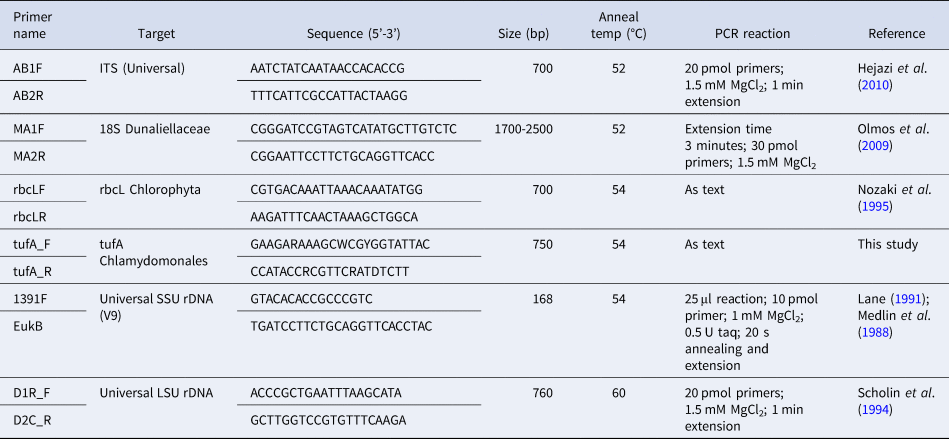
Table 2. Primers used in this study
Sequences were manually verified for quality using Chromas (Technelysium Pty Ltd). Multiple sequence alignments were constructed in BioEdit 7.0 (Hall, Reference Hall1999) using ClustalW. Phylogenetic analysis based on neighbour-joining and maximum likelihood was undertaken using MEGA 6 (Tamura et al., Reference Tamura, Stecher, Peterson, Filipski and Kumar2013). Bootstrap values were retrieved from 1000 replicates. Accession numbers for sequences generated in this study are detailed in Supplementary Table 1.
Results
Microscopy
Microscopy clearly identified cells that had accumulated β-carotene owing to the orange-red colouration and hence those affiliated to the section Dunaliella, species D. salina (Figure 1). These cultures (DF40, DF41, DF45, DF17, DF15, T36, T27, T41, T68, SA4 and SA3) ranged in mean cell length of 12.6–18.2 μm and cell width of 10.2–15.1 μm (Table 1). Cell shape was variable within cultures and stigma was not easily identifiable due to the presence of refractile granules.
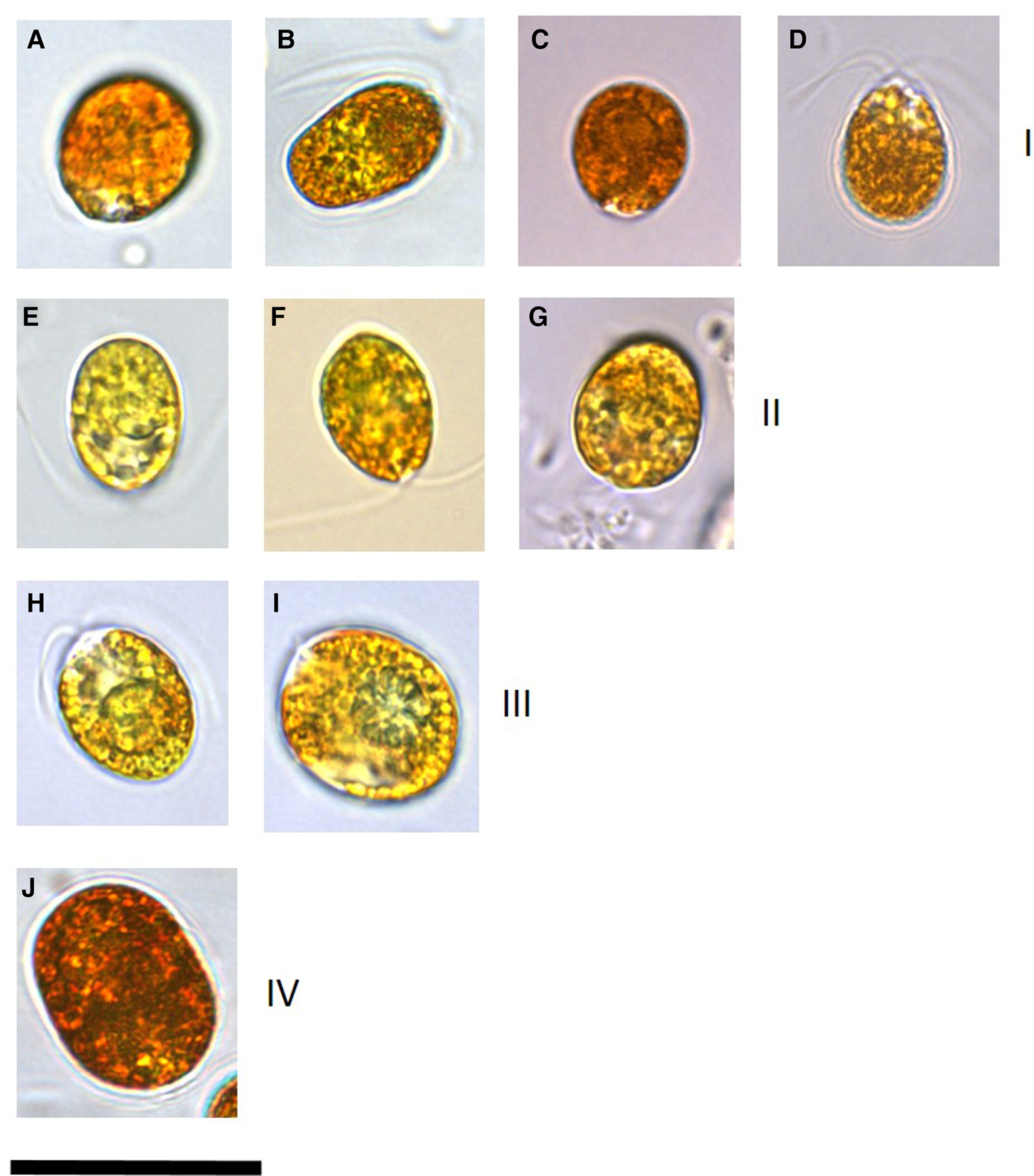
Fig. 1. Images of a representative set of Dunaliella salina strains isolated in this study compared with strains imaged from culture collections. All isolates were cultured in identical conditions and imaging for each isolate was undertaken 2 weeks following sub-culturing. Numerals in brackets after the strain name indicates the D. salina sub-group. (a) D. salina DF40 (I), (b) D. salina DF45 (I), (c) D. salina CCAP 19/18 (I), (d) D. salina MUR22 (I), (e) D. salina DF17 (II), (f) D. salina CCAP 19/25 (II), (g) D. salina UTEX 1644 (II), (h) D. salina T41 (III), (i) D. salina T37 (III), (j) D. salina DF15 (IV). Scale bar is equivalent to 25 μm.
18S intron-sizing method and sequence analysis
As anticipated, according to Olmos et al. (Reference Olmos, Ochoa, Paniagua-Michel and Contreras2009), D. tertiolecta had no introns (Table 3). We also identified two different groups of D. salina, as reported in Olmos et al. (Reference Olmos, Ochoa, Paniagua-Michel and Contreras2009), those with two introns and those with one. Further to this, however, some isolates that were assigned as D. salina microscopically were found to have no introns, i.e. they had the same MA1/2 18S amplicon size as D. tertiolecta. In this study, we PCR-amplified from two of the same isolates as Olmos and co-workers, with differing results. They reported D. bardawil UTEX 2538 to have two introns whereas we sized it to have one. Moreover, we sized D. bardawil ATCC (American Type Culture Collection) 30861, which is thought to be the same strain as UTEX 2538, and found it to have no introns. Strain ATCC 30861 was isolated by Ben-Amotz and Avron in 1976 from a salt pond near Bardawil, Israel, and this strain was then deposited in the UTEX collection as strain 2538 by R. Adams between 1980 and 1982. The culture collection legacy of this strain continues as two other culture collections host this strain as the same as the original D. bardawil isolated by Ben-Amotz and Avron. SAG (Culture Collection of Algae at Göettingen University) received the strain from Avron via Prof Thompson in 1988, re-naming it D. salina SAG 42.88 in 2001, and CCAP (Culture Collection of Algae and Protozoa) received the strain from SAG in 1996 (Muller, Reference Muller2005) identifying it as strain 19/30. Olmos et al. (Reference Olmos, Ochoa, Paniagua-Michel and Contreras2009) reported the CCAP 19/30 sequence in GenBank, that was deposited by ‘Herve’ in 2006, to have one intron which is in agreement with our result for ATCC 30861, however, both our study and Olmos et al. (Reference Olmos, Ochoa, Paniagua-Michel and Contreras2009) reported CCAP 19/30 to have no introns. In this study we also sequenced the 18S amplicons for the different strains and for those strains that had introns all of them shared the same sequence with the exception of DF15 (Table 3).
Table 3. Results from intron sizing and sequence alignment of introns
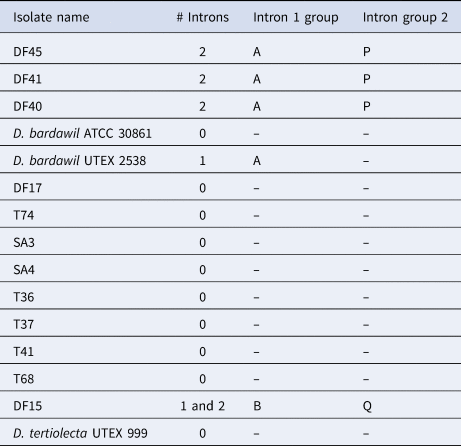
DF15 was the only strain to produce two different-sized amplicons of the 18S rDNA with the intron-sizing method, leading us to suspect contamination of the culture. To confirm this was not a result of contamination, further rounds of single-cell picking from the culture were undertaken to ensure clonality and six more cells were picked from this culture, washed and grown in higher salt media (final concentration 185 g L−1 sea salts) to ensure selection for the halophilic D. salina. The 18S PCR was repeated with the six newly isolated clones with the PCR consistently generating two amplicons. Both PCR products were sequenced, with the smaller product found to contain one intron and the larger product with two. When the sequences were compared, nucleotide substitutions were detected across the whole sequence (both within the intron and exon regions). For the intron that the two DF15 amplicons shared, there was 98.5% similarity and the large amplicon intron and small amplicon intron shared 89.6% and 88.8% identity with D. bardawil UTEX 2538, respectively. BLASTn analysis showed these introns to be most similar to Dunaliella sp. ST13 (>98%). The second intron in the larger product was unique, sharing 99.23% identity also with Dunaliella sp. strain ST13.
Phylogenetic analysis
Strains CCAP 19/30 and MUR 8 were omitted from the phylogenetic analysis as morphological analysis showed that they did not turn red under stress. Furthermore, BLAST analysis of the sequences generated from these two strains and also CONC 001 showed them to be more similar to D. tertiolecta (data not shown) and hence confirmed their exclusion from the D. salina phylogeny. GenBank holds a vast number of sequences originating from Dunaliella species which are available to include in phylogenetic analysis. Because of some of the problems reported with strain identification and/or cross contamination we chose to only include key strains that we obtained from culture collections or DNA where the culture was not available.
The large-subunit (LSU) rDNA phylogeny formed a strongly supported clade of five out of seven of the D. salina strains isolated from Salar de Atacama, Chile, in 1990, however, other subclades were weakly supported, owing to only a small number of nucleotide substitutions (Figure 2). The clade containing D. bardawil UTEX 2538 clustered separately from D. salina UTEX 1644 owing to 6 bp substitutions across 477 bp of the alignment.
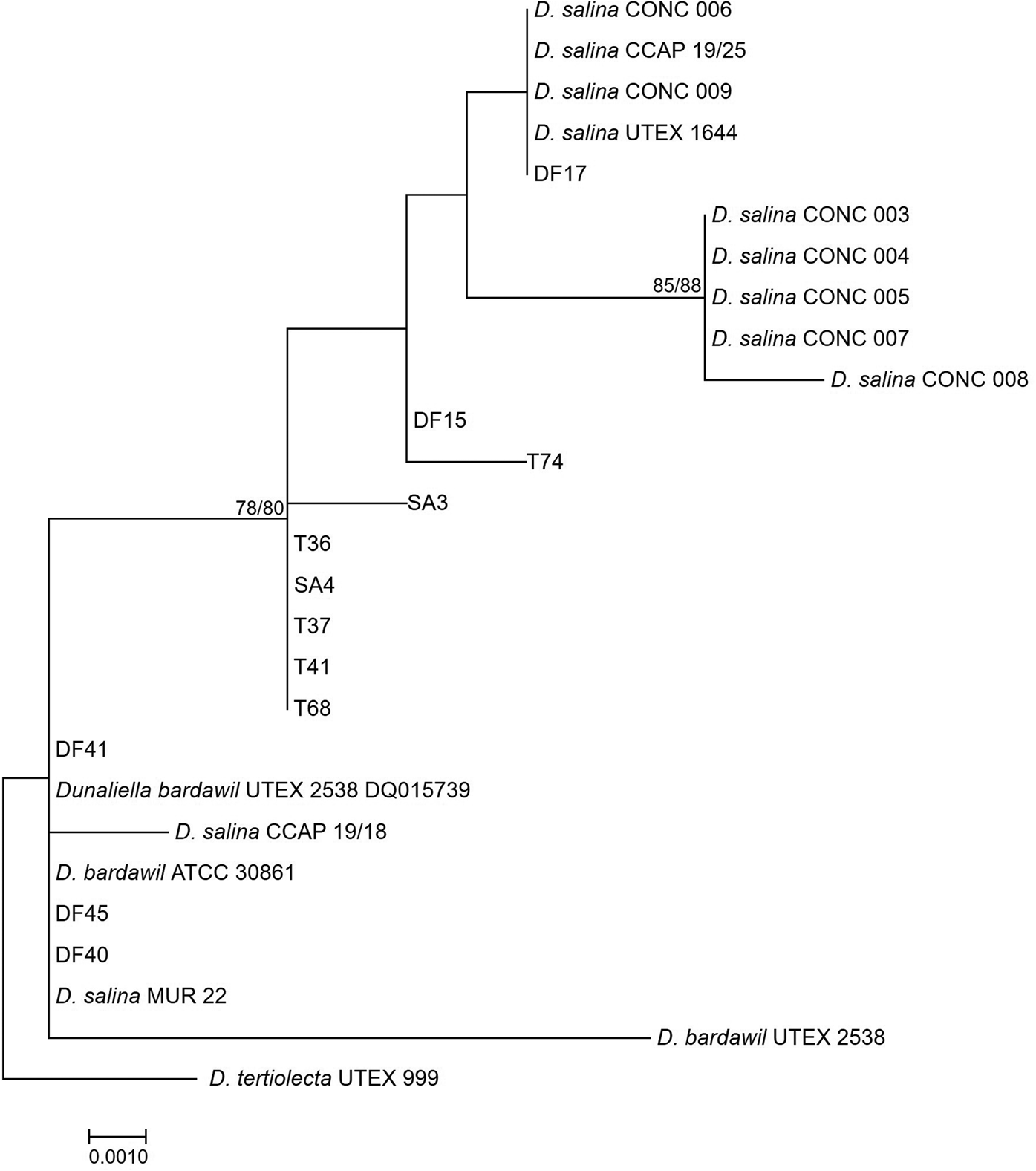
Fig. 2. Neighbour-joining tree of D. salina strains isolated during this study and sequences from GenBank based on a 477 bp alignment of the LSU gene. Bootstrap values were retrieved from 1000 replicates and those >70% are indicated at the nodes for neighbour-joining and maximum likelihood respectively. The out-group was D. tertiolecta UTEX 999.
The rbcL phylogeny (Figure 3) divided D. salina broadly into two separate clades, corresponding to the clustering patterns described by Assunção et al. (Reference Assunção, Jaén-Molina, Caujapé-Castells, de la Jara, Carmona, Freijanes and Mendoza2012) (salina I and salina II). The rbcL phylogeny further sub-divides the Chilean clade identified in the LSU phylogeny, but with the addition of CONC 009 which clustered separately in the LSU phylogeny.
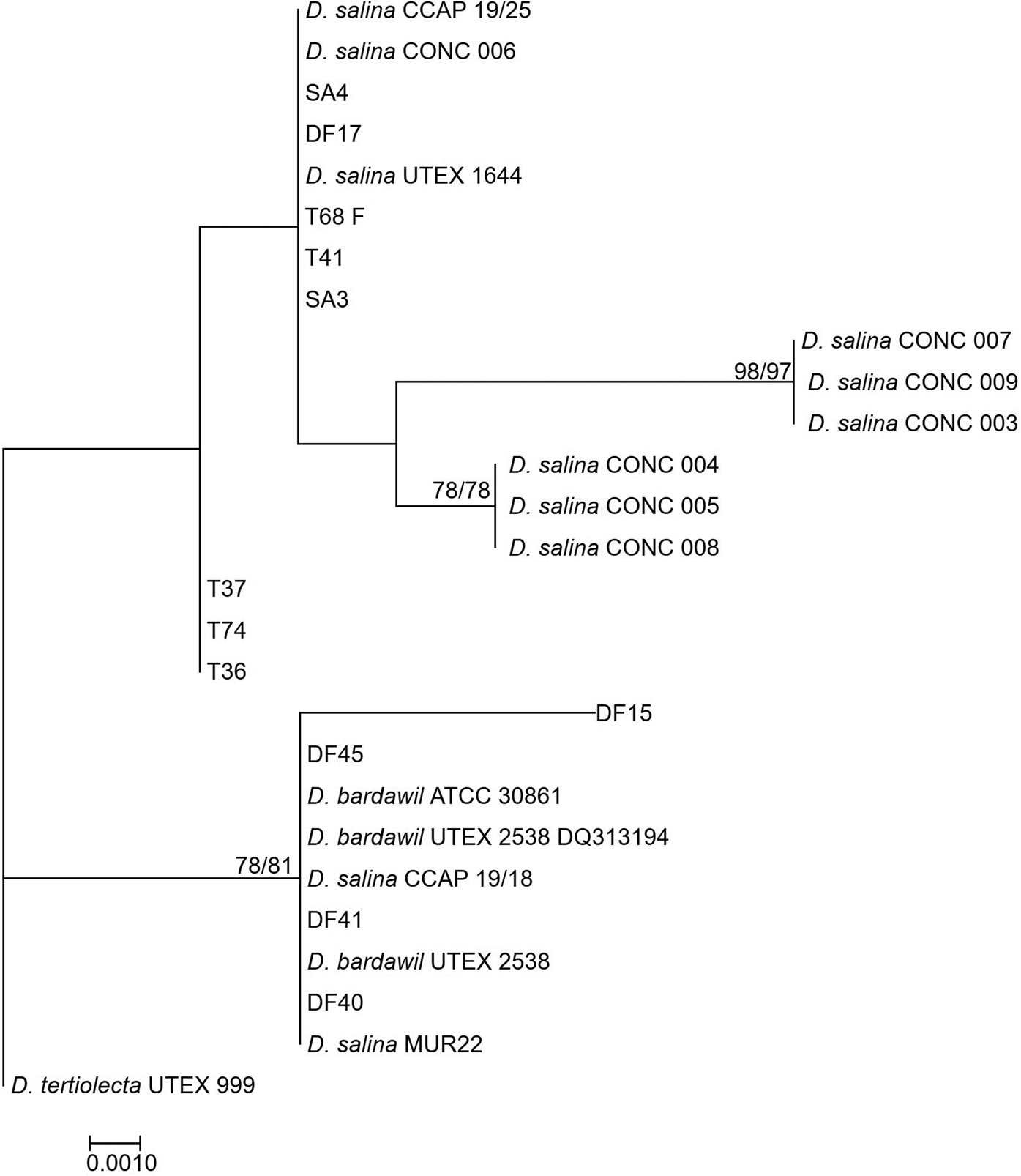
Fig. 3. Neighbour-joining tree of D. salina strains isolated during this study and sequences from GenBank based on a 521 bp alignment of the rbcL gene. Bootstrap values were retrieved from 1000 replicates and those >70% are indicated at the nodes for neighbour-joining and maximum likelihood respectively. The out-group was D. tertiolecta UTEX 999.
The tufA phylogeny (Figure 4) grouped all the new isolates and previously characterized D. salina strains in one clade with all the sequences >99% similar to each other.

Fig. 4. Neighbour-joining tree of D. salina strains isolated during this study and sequences from GenBank based on a 614 bp alignment of the tufA gene. Bootstrap values were retrieved from 1000 replicates and those >70% are indicated at the nodes for neighbour-joining and maximum likelihood respectively. The out-group was D. tertiolecta UTEX 999.
The phylogenetic tree based on ITS1 + ITS2 (Figure 5) provided much more resolution compared with the other trees and was robust, supported by high bootstrap values. In order to complement the phylogeny of González et al. (Reference González, Gómez, Polle, Ben-Amotz, Polle and Subba Rao2009) who identified three main D. salina clades, we included the sequence for D. salina AC144 in this analysis. This strain was the sole member of one of the clades identified by González et al. (Reference González, Gómez, Polle, Ben-Amotz, Polle and Subba Rao2009) and therefore was of interest to examine its relationship with the strains from this study. Two salina groups were identified in this phylogeny, according to Assunção et al. (Reference Assunção, Jaén-Molina, Caujapé-Castells, de la Jara, Carmona, Freijanes and Mendoza2012), and these were clearly separated by long branch lengths with salina I which encompasses D. bardawil UTEX 2538 and salina II which encompasses D. salina UTEX 1644. The Chilean strains were separated into two groups, matching the LSU phylogeny. Due to ambiguous bases in the DF15 sequences, this amplicon was cloned and sequenced, hence the inclusion in the tree of four DF15 sequences. DF15 clones were found to have nucleotide substitutions across the ITS region when sequenced with the three sequences identified found to share >99% identity. In the LSU phylogeny (Figure 2) DF15 clustered with the D. salina UTEX 1644 (salina II) group but with long branch lengths separating them. However in the rbcL (Figure 3) phylogeny, this strain groups with D. bardawil UTEX 2538 (salina I). The ITS phylogenetic tree places DF15 intermediary to the clades classified as salina I or II by Assunção et al. (Reference Assunção, Jaén-Molina, Caujapé-Castells, de la Jara, Carmona, Freijanes and Mendoza2012). BLASTn analysis of the DF15 ITS sequence identified Dunaliella sp. ST13 as most similar yet with only 98.67% identity. The sequence for D. salina AC144 weakly clustered with the top half of the tree with low bootstrap values.

Fig. 5. Neighbour-joining tree of D. salina strains isolated during this study and sequences from GenBank based on a 378 bp alignment of the ITS1 + ITS2. Bootstrap values were retrieved from 1000 replicates and those >70% are indicated at the nodes for neighbour-joining and maximum likelihood respectively. The out-group was D. tertiolecta UTEX 999.
The concatenated tree of the 4 markers, ITS-LSU-rbcL-tufA (Figure 6) was constructed to further resolve/affirm any clades/sub-clades to provide reliable information on the taxonomy of these strains in combination with microscopy. Four sub-clades of Dunaliella salina could be identified, and these were supported by significant bootstrap values. Within each of the subclades the strains shared 99% identity, with all the strains morphologically identified as D. salina sharing at least 97% identity.

Fig. 6. Neighbour-joining tree of D. salina strains isolated during this study and sequences from GenBank based on an alignment of a concatenation of the ITS-LSU-rbcL-tufA sequences used to produce Figures 2–5. Bootstrap values were retrieved from 1000 replicates and those >70% are indicated at the nodes for neighbour-joining and maximum likelihood respectively. Images show the morphology of representative strains. The out-group was D. tertiolecta UTEX 999.
V9 (SSU rDNA) amplicon sequencing was undertaken for DF15, DF17, D. bardawil UTEX 2538, D. salina CONC 008, D. salina CCAP 19/25, D. salina UTEX 1644, D. salina CCAP 19/18 and D. tertiolecta UTEX 999 only (Figure 7). Nucleic acid alignment showed that Dunaliella of the tentative clades, salina I, and tertiolecta were identical, however salina II, was different. DF15 was identical to salina I and D. tertiolecta.
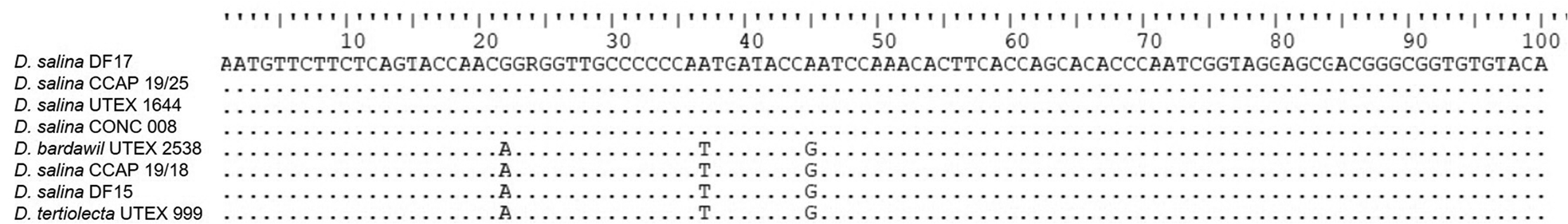
Fig. 7. CLUSTALW alignment of the V9 SSU sequences generated in this study and selected sequences from GenBank. Dots indicate identical nucleotides and letters indicate nucleotide substitutions.
Discussion
The intron sizing method previously revealed useful information regarding the history of Dunaliella spp. in culture collections. Whilst providing some data indicating the taxonomic affiliation of particular strains we found the method was not sufficiently robust. The fact that a group of D. salina have no introns (the same as D. tertiolecta) negates the intron-sizing as a tool for separating these strains from each other as both an isolate of D. tertiolecta and D. salina would produce the same size amplicon, despite these two being very different species. Selection based on biochemistry could however easily separate these two species prior to intron-sizing if this method was preferred. Olmos et al. (Reference Olmos, Ochoa, Paniagua-Michel and Contreras2009) concluded that cross-contamination of cultures had occurred which resulted in differences in amplicon sizes in what was thought to be the same strain. Our study showed similar discrepancies with identical strains D. bardawil, strain UTEX 2538 and strain ATCC 30861 from separate culture collections having a different number of introns. This could indicate cross-contamination in its culturing history, primary non-clonal cultures or could indicate intron loss in one of the strains in the different culture collections. Indeed in our phylogenetic analyses the two aforementioned strains consistently group together with identical tufA and rbcL sequences, ITS sequences differing by 2 bp over a 553 bp multiple sequence alignment and LSU sequences differing by 5 bp substitutions over 477 bp. Considering the original isolate was collected in 1976, genetic divergence in separate culture collections is conceivable although without more information this cannot be determined. Certainly, with advances in cryopreservation one would be able to track such events more closely by resurrecting and comparing with regularly sub-cultured algae.
Our study confirms that, based on the sequencing analysis of four marker genes, the strains CCAP 19/30, MUR 8 and CONC 001 that we acquired in this study are not D. salina. This is not to say that other cultures of these strains around the world are not true representatives of the original designation of these strains, however, but because of our observations they were not included in the analyses here. These results are a cautionary tale for anyone working on algal cultures, with genotyping of the strains recommended as an integral step to ascertain the taxonomic affiliation of the strains one is studying.
The sequences for DF15 nuclear SSU rDNA were shown to be unique, with two different versions amplified. Whilst there is a high copy number of the rDNA, concerted evolution typically moves towards resulting identical sequences of this gene within the genome so it is surprising to get two different versions of this gene in DF15. Since its clonality was verified through multiple rounds of single cell picking, intragenomic sequence variation is a possible explanation. Certainly, Alverson & Kolnick (Reference Alverson and Kolnick2005) reported intragenomic nucleotide polymorphisms within the SSU rDNA of the diatom Skeletonema, however, the differences identified were random single nucleotide polymorphisms rather than whole introns. We propose that DF15 represents a novel lineage and as such concerted evolution has not progressed to completion as seen in the other Dunaliella species. Alternatively, it is possible that the rate of variation is exceeding concerted evolution with the hypersaline harsh environment selecting for heterogeneity. However, if this was indeed the case one would anticipate identifying this heterogeneity in more strains.
Comparing the four markers analysed in this study shows that tufA is the least useful in resolving intraspecific diversity within D. salina. However, the tufA analysis has been of use in that it supports the designation of all of these strains to the species D. salina. González et al. (Reference González, Gómez, Polle, Ben-Amotz, Polle and Subba Rao2009) discussed the high intraspecific diversity within D. salina and the potential existence of more than one biological species, yet these data support the existence of only one. The LSU and rbcL phylogenies show more resolution than tufA, providing some information on the relationships between the different strains, but are less robust than the ITS. The phylogenetic tree based on the ITS regions gives the most information, with subdivisions strongly supported by high bootstrap values. The concatenated tree of the four genes further reinforces the ITS region phylogeny with a clearer delineation of different phylogroups. Including intron presence/absence data and sequences can provide further supporting information on some subdivisions, such as the separate DF15 clade.
As a marker on its own, the V9 region of SSU rDNA does not offer enough variability for accurate taxonomy for the D. salina species. The V9 region lends itself to next generation sequencing methodologies, due to its heterogeneity and short length, however, the fact that not all the groups can be resolved with this marker raises questions on its suitability for analysis of hypersaline microbial communities, as a significant portion of D. salina diversity will be missed. The V2–V4 region of the SSU rDNA was found to have the best phylogenetic resolution compared with the other hypervariable regions in dinoflagellates (Ki, Reference Ki2012), however, this was not tested here.
There is definitive D. salina taxa defined by its morphology and physiology with sequence information from our study demonstrating a robust segregation into four clades. We support the two sub-clades designated by Assunção et al. (Reference Assunção, Jaén-Molina, Caujapé-Castells, de la Jara, Carmona, Freijanes and Mendoza2012), salina I and II, and identify two further groups. Salina II, as previously identified (Assunção et al., Reference Assunção, Jaén-Molina, Caujapé-Castells, de la Jara, Carmona, Freijanes and Mendoza2012), comprises the original isolate of what we know as D. salina, isolated in 1967 by Loeblich from Baja California and is deposited within the UTEX culture collection as #1644. The salina I clade contains originally classified D. bardawil strains including the first isolate within this clade, D. bardawil UTEX 2538 isolated in 1976 and also D. salina CCAP 19/18, which has recently been proposed as a reference strain for the species by Polle et al. (Reference Polle, Jin and Ben-Amotz2020). We propose two other sub-clades within D. salina, the salina III group which encompasses strains T36, T37, T41, T68, SA3 and SA4 isolated in this study. These form a robust group and therefore are considered distinct from the salina II group. Finally, we propose a new salina IV sub-clade that contains DF15 as the sole member. The ITS phylogeny potentially also suggests the D. salina AC144 (as analysed by González et al., Reference González, Gómez, Polle, Ben-Amotz, Polle and Subba Rao2009) is a unique strain, however due to unavailability of this strain, comprehensive genotyping could not be undertaken to corroborate this.
Strain DF15 is a distinctive strain of D. salina as, significantly, it does not delineate with either group. This strain is somewhat of an enigma with the different markers used clustering it (weakly) with different groups, e.g. DF15 clusters with strain DF17 for LSU but with strain UTEX 2538 in the rbcL phylogenies. Interestingly, biochemical analyses of this strain found it to be the highest producer of all-trans β-carotene, 9-cis β-carotene and zeaxanthin compared with four other D. salina strains, which included DF17, DF40, CCAP 19/30 and UTEX 2538, when tested at 1500 μmolm−2 s−1 light intensity (Xu et al., Reference Xu, Ibrahim, Wosu, Ben-Amotz and Harvey2018). We therefore propose that the salina IV lineage we have identified may comprise of highly sought after strains for biochemical exploitation.
However, principal component analysis performed by Xu et al. (Reference Xu, Ibrahim, Wosu, Ben-Amotz and Harvey2018) examined 11 traits for DF15, DF17, DF40 and UTEX 2538 (all-trans β-carotene, 9-cis β-carotene, glycerol, lutein, zeaxanthin, all-trans α-carotene, photosynthesis, respiration, total carotenoids, total chlorophyll and specific growth rate at four different light intensities) and showed that DF17 and DF40 clustered closely whereas our phylogenetic analysis separates them into different clades. This highlights the ambiguities between genetic data and biochemical data, however, we believe that these data are supportive of DF15 belonging to a unique sub-clade of D. salina. Upon identifying other strains that can be assigned to this clade we can confirm that it is a group characterized by the production of exceptional quantities of carotenoids. Genetic information from this strain and the identification of its sub-clade could be used to screen environments for this strain which have the potential to produce similarly high quantities. Due to ongoing revisions and changes in D. salina taxonomy we have refrained from the designation of new sub-species.
In conclusion this study has revealed the further diversity of D. salina isolates collected from South Africa, Israel, Namibia, Spain and in particular the novel strain DF15. In combination with morphological analysis the suite of genes used for molecular analysis has permitted the separation of D. salina into four sub-clades. Certainly, it appears to be beneficial to use several gene markers for phylogeny in order to generate a robust tree, however at the very least we are in agreement with other studies that the ITS regions are the most appropriate for resolving different clades. However, we propose that ITS2 should be used with ITS1 and 5.8S to provide even more genetic information. The classification of Dunaliella has proven to be a challenge over many decades and we recommend that the Dunaliella community needs to come together to tackle the challenge in unison with the vision to facilitating the selection of important strains for key uses.
Supplementary material
The supplementary material for this article can be found at https://doi.org/10.1017/S0025315420001319
Acknowledgements
We thank Patricia Gomez for supplying DNA for Chilean Dunaliella sp. and Prof. W. Wilson for helpful comments that improved the manuscript. We would also like to thank Dr Maya Pfaff, Department of Environmental Affairs, Forestry and Fisheries South Africa for assisting Prof. Declan Schroeder in the sampling of the South African salt pans and Sophia Rhodes for providing her facilities for purposes of sample processing prior to transport.
Financial support
This research was supported by the D-factory and the EU KBBE.2013.3.2-02 programme (D-Factory: 368 613870) and the ValgOrize consortium, subsidised by the Interreg 2 Seas programme 2014–2020, co-financed by the European Fund for Regional Development in the framework of subsidy contract nr. 2S05-17.












