Introduction
Malarial parasites (Phylum: Apicomplexa, Order: Haemosporida) are eukaryotic protozoan parasites that infect a diversity of vertebrate hosts such as birds, squamate lizards and mammals (Garnham, Reference Garnham1966; Perkins and Schaer, Reference Perkins and Schaer2016). Bats, with over 1349 species (Simmons, Reference Simmons, Wilson and Reeder2005; Miller-Butterworth et al., Reference Miller-Butterworth, Murphy, O'Brien, Jacobs, Springer and Teeling2007; Lack et al., Reference Lack, Roehrs, Stanley, Ruedi and Van Den Bussche2010) are mammalian hosts to a large diversity of haemosporidian parasites (e.g. Schaer et al., Reference Schaer, Perkins, Decher, Leendertz, Fahr, Weber and Matuschewski2013). Phylogenetic analyses suggest that bats were likely the first mammals to acquire infections with haemosporidians before subsequent transmission to other non-chiropteran mammals (Schaer et al., Reference Schaer, Perkins, Decher, Leendertz, Fahr, Weber and Matuschewski2013; Lutz et al., Reference Lutz, Patterson, Kerbis Peterhans, Stanley, Webala, Gnoske, Hackett and Stanhope2016). Bats are parasitized by nine different haemosporidian genera, seven of which are exclusive to them (Perkins and Schaer, Reference Perkins and Schaer2016). The haemosporidian genera that infect bats include Plasmodium, Hepatocystis, Polychromophilus and Nycteria with each of these being host-specific to particular bat families within the order Chiroptera (Schaer et al., Reference Schaer, Perkins, Decher, Leendertz, Fahr, Weber and Matuschewski2013). Chiropteran haemosporidians are therefore important for the study of the evolution of malaria parasites (Perkins and Schaer, Reference Perkins and Schaer2016). Gaining insights on the ecology and evolution of bat malaria parasites and its potential impacts on the hosts systems will guide future conservation efforts and advance studies on the impacts parasites have on hosts.
Nigeria is a biodiversity hotspot for bats with approximately 71 described species distributed within the country (IUCN, 2017). Here, we investigated the prevalence and phylogenetic relationships of haemosporidian parasites among bats in Nigeria.
Materials and methods
Bats were sampled in the Amurum Forest Reserve (9°53′N, 8°59′E) which is located in Laminga village, 15 km northeast of Jos, Plateau state, North-Central Nigeria during May and June in 2015 and 2016 at twelve different sites (Fig. 1A, Table S1). Bats were captured opportunistically using ground-level mist nets and canopy nets and kept individually in cloth bags prior to sampling. Standard measurements including sex, age and reproductive condition were recorded and species identified using the key of Rosevear (Reference Rosevear1965). The responsible institution for the approval of the use of Amurum Forest Reserve for research, the Scientific and Research Ethics Committee of the A. P. Leventis Ornithological Research Institute (APLORI), reviewed and officially approved this survey (4 May 2015).
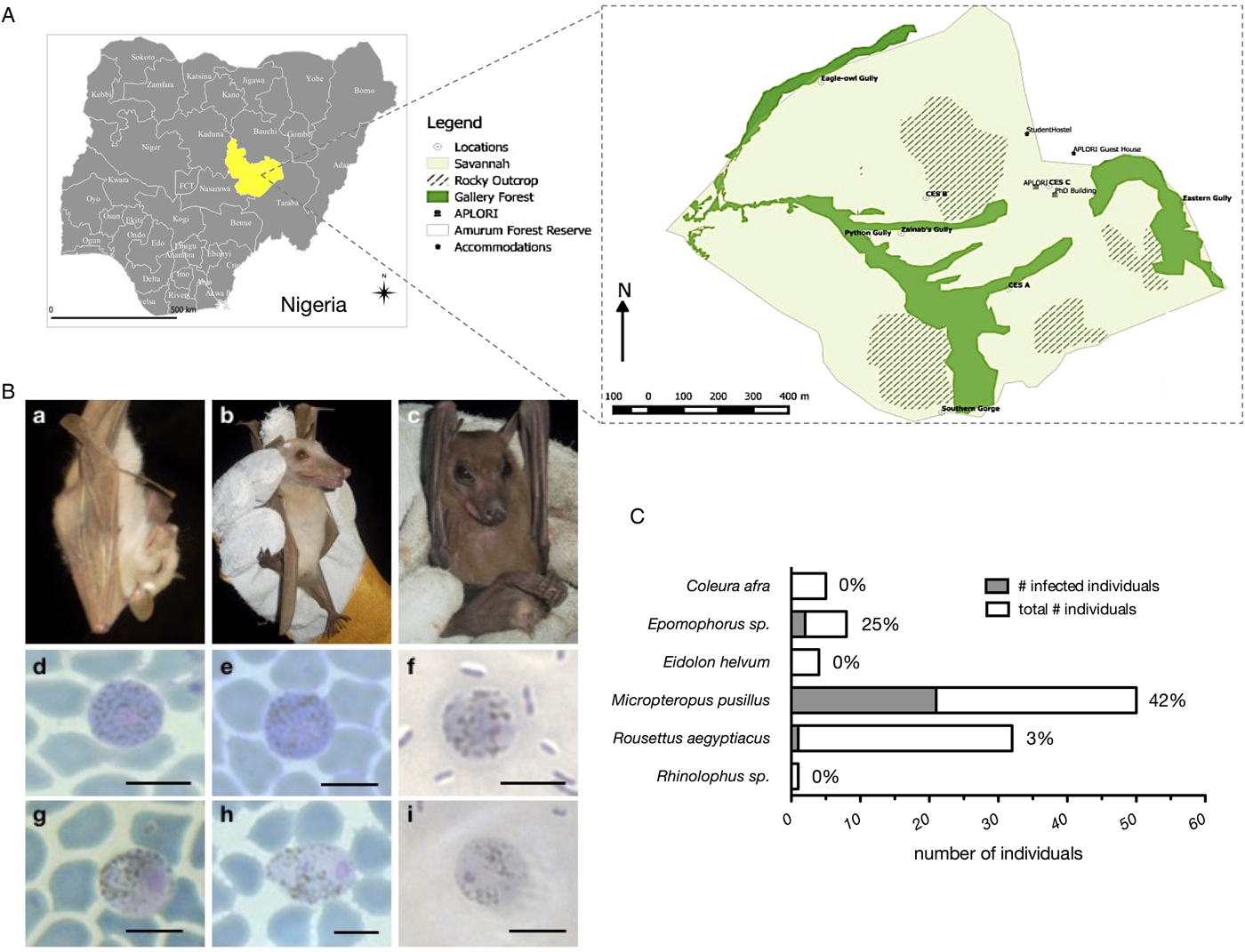
Fig. 1. (A) Amurum Forest Reserve in Jos plateau state Nigeria. The site covers an area of about 300 hectares with a mean annual rainfall of 1375–1750 mm per annum and a mean temperature of 10 °C during the cold season (December–February) and 32 °C during the hot season (April–May). The vegetation is a rocky outcrop in dry scrub savannah with gallery forests and patches of grassland. Bats were sampled at 12 sites, which cover all three habitat types, Savannah (light green), gallery forests (dark green), and rocky outcrops (striped), during May/June of 2015 and 2016. The maps were generated with QGIS. (B) Host species and representative micrographs of Hepatocystis blood stages. Shown are examples of the infected pteropodid bats, Micropteropus pusillus (a), Epomophorus sp. (b), and Rousettus aegyptiacus (c). Micrographs depict macrogametocytes (d = FB50, M. pusillus; e = FB31, M. pusillus; f = FB12, R. aegyptiacus) and microgametocytes (g = FB51, M. pusillus; h = FB52, M. pusillus; I = FB12, R. aegyptiacus). Only mature stages of gametocytes were detected, featuring bright-blue (macrogametocytes, d, e, f) or biscuit-coloured (microgametocytes, g, h, i) cytoplasm and coarse pigments as described for Hepatocystis in other African epauletted bats (e.g. Garnham, Reference Garnham1966). The nucleus of the microgametocytes was characteristically surrounded by a pigment-free area. Magnification is 1000×. Size bars = 5 µm (C) Prevalence of haemosporidian infections in %.
Blood samples were collected by pricking the cephalic vein with a sterile 27G needle and collecting approximately 100 µL of blood into heparinized capillary tubes (samples from 2015) or dotted on FTA® DNA cards (samples from 2016). Blood from the capillary tubes was immediately used to prepare thin blood smears, which were air-dried and fixed in 99–100% (vol/vol) ethanol for five minutes. Sampled bats were marked with commercial nail polish on their hind claws to avoid short-term re-sampling and were released at the capture site.
Blood smears were re-fixed with 99–100% (vol/vol) ethanol or methanol prior to Giemsa staining. Screening of stained blood smears and morphological identification of haemosporidian parasites were performed using light microscopy (Nikon Instrument Inc., USA) under oil immersion at 1000× magnification (Fig. 1B).
DNA was extracted from unstained blood smears on glass slides that was removed using a sterile razor blade (2015 samples) or from FTA cards (2016 samples). All DNAs were extracted using the Qiagen DNeasy extraction kit (Hilden, Germany), using products protocol for animal tissues with elution volumes of 30 and 60 µL for the 2015 and 2016 blood samples respectively. PCRs were performed using the QIAGEN TopTaq Master Mix. The mitochondrial gene cytochrome b (cytb) was amplified with the primer sets DW2/3932R and Hep-F3/Hep-R3 and the nuclear elongation factor 2 gene (ef2) using the primers EF2-F/EF2-R (Schaer et al., Reference Schaer, Perkins, Decher, Leendertz, Fahr, Weber and Matuschewski2013). The apicoplast caseinolytic protease (clpC) was amplified using nested PCR (Martinsen et al., Reference Martinsen, Perkins and Schall2008) (Table S2). All PCR products were sequenced with the amplification primers in both directions using BigDye v3.0 (Applied Biosystems) and run on an ABI-373 sequencer. In order to verify morphological bat species identifications, bat genetic markers comprising nuclear introns and protein-coding genes (acox2, fgb, rogdi, rag1, rag2) were amplified and sequenced (Tables S2 and S3).
Parasite and bat DNA sequences were edited using Geneious version 8.1.9 software, with ambiguous positions or missing data coded with N′ and sequences aligned using the Muscle algorithm (Edgar et al., Reference Edgar, Drive and Valley2004). Parasite sequences included 906 nucleotides (nt) of cytb, 528 nt of clpC and 516 nt of the nuclear ef2-gene (Table S3). Reference sequences, including the major mammalian haemosporidian clades and Hepatocystis sequences from primates and African bats were retrieved from GenBank and added to the alignments (Table S3). Sequence alignments of the three genes cytb, clpC and ef2 were concatenated and phylogenetic relationships were evaluated with Bayesian inference methods using Leucocytozoon as outgroup taxon. Data were divided into partitions according to the three genes and PartitionFinder vs.2 was used to test different DNA substitution models and partition schemes (Lanfear et al., Reference Lanfear, Frandsen, Wright, Senfeld and Calcott2017). Bayesian inference was conducted in MrBayes v3.2.6 (Huelsenbeck and Ronquist, Reference Huelsenbeck and Ronquist2001), via the CIPRES Science Gateway Web Portal V3.3 (Miller et al., Reference Miller, Pfeiffer and Schwartz2010) with two runs of four chains (three heated, one cold, temperature = 0.03) each for 10 million generations. The first 25% of trees were discarded as burn-in. Tracer v1.6 (Rambaut et al., Reference Rambaut, Suchard, Xie and Drummond2014) was used to assess mixing and convergence of runs and effective sample size (ESS > 500). Phylogenetic trees were visualized with the program FigTree (http://tree.bio.ed.ac.uk/software/fgtree/).
Results
A total of 100 bats of six species belonging to three families were caught at nine of the twelve sites within Amurum Forest Reserve (Fig. 1A, Tables S1 and S4). The majority of the bats (95%) belonged to the fruit bat family Pteropodidae, with the four species: Epomophorus sp., Micropteropus pusillus, Eidolon helvum and Rousettus aegyptiacus. In addition, a single Rhinolophus sp. (Rhinolophidae) and five Coleura afra (Emballonuridae) were captured. The overall prevalence of haemosporidian parasites was 25% (25/100) with parasites exclusively detected in the bat family Pteropodidae (Fig. 1C, Table S4). The two epauletted fruit bat genera Micropteropus and Epomophorus had a parasite prevalence of 42% (21/50) and 25% (2/8), respectively. We further detected a haemosporidian infection in the single R. aegyptiacus (3%, 1/32) that was sampled.
The morphology of the gametocyte blood stages of the parasites corresponds to the descriptions of Hepatocystis parasites of epauletted fruit bats from Guinea, Liberia, Cote d'Ivoire and South Sudan (Schaer et al., Reference Schaer, Perkins, Decher, Leendertz, Fahr, Weber and Matuschewski2013, Reference Schaer, Perkins, Ejotre, Vodzak, Matuschewski and Reeder2017) (Fig. 1B). The phylogenetic analysis confirmed all parasites as belonging to the genus Hepatocystis. Sequences of the samples of the study closely group within the Hepatocystis parasite clades of epauletted fruit bat species from different African countries (Fig. 2). No host species specificity is apparent, which is in agreement with previous findings of Hepatocystis in bats (Schaer et al., Reference Schaer, Perkins, Ejotre, Vodzak, Matuschewski and Reeder2017). The analyses confirmed Hepatocystis as monophyletic clade (posterior probability of 1) with the African bat Hepatocystis sequences grouping in one monophyletic subclade as previous studies have shown (Fig. 2) (Schaer et al., Reference Schaer, Perkins, Ejotre, Vodzak, Matuschewski and Reeder2017). The Hepatocystis sequences of the infected R. aegyptiacus individual group among the parasite sequences of the epauletted fruit bats. However, compared to all other Nigerian samples of the study, the parasite sequences of R. aegyptiacus slightly differ (e.g. 28 unique bases across 890 nt of the cytb sequence, representing a 3% sequence divergence) and do not group with the other Nigerian sequences, but with sequences from parasites of Myonycteris from Cote d'Ivoire and diverse epauletted hosts from South Sudan (Fig. 2).
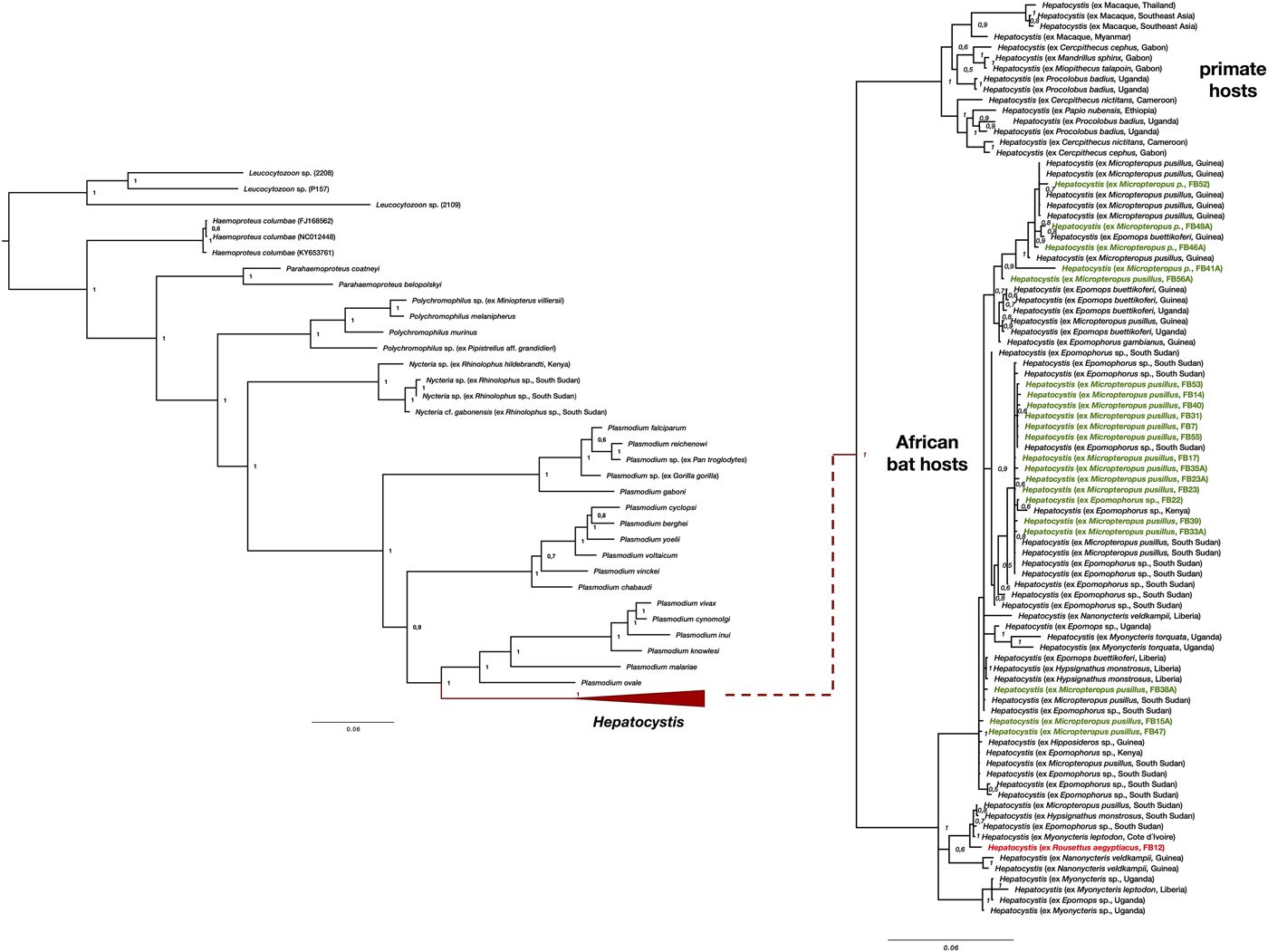
Fig. 2. Phylogenetic analysis of Hepatocystis parasites in the context of the major haemosporidian clades recovered by Bayesian analysis. Posterior probabilities are given. The analysis was performed on the concatenated alignment (total of 1947 bp) of the mitochondrial cytochrome b (906 bp), the apicoplast caseinolytic protease (538 bp), and the nuclear elongation factor 2 (516 bp). (A) Placement of Hepatocystis as collapsed clade as sister to the mammalian Plasmodium (Plasmodium) clade comprising Plasmodium vivax and Plasmodium ovale, as shown before (e.g. Schaer et al., Reference Schaer, Perkins, Decher, Leendertz, Fahr, Weber and Matuschewski2013). (B) The uncollapsed clade contains representative reference sequences of Hepatocystis of primate hosts as well as African bat hosts. The samples of the current study are highlighted in bold (parasite sequences from Epomophorus and Micropteropus hosts in green, the sequence from Rousettus in red). Sequences of the Nigerian samples closely group within the Hepatocystis parasite clades of epauletted fruit bat species from different African countries and no host species specificity is apparent. The Hepatocystis sequences of R. aegyptiacus group among the parasite sequences of epauletted fruit bat species from South Sudan and with sequences from parasites of Myonycteris from Cote d'Ivoire and slightly differ from the Nigerian Epomophorus and Micropteropus parasite sequences.
Discussion
This study provides the first information on the haemosporidian parasites of bats in Nigeria through an investigation of bat malaria parasites in the Amurum Forest Reserve. Hepatocystis was discovered in epauletted fruit bats in Nigeria and the study strengthens the notion that the bat genera Epomophorus and Micropteropus present common hosts of Hepatocystis parasites in several African countries. The prevalences (of 25 and 42%) are similar to previous reports from epauletted fruit bats in other West, Central and East African countries (e.g. Schaer et al., Reference Schaer, Perkins, Decher, Leendertz, Fahr, Weber and Matuschewski2013; Lutz et al., Reference Lutz, Patterson, Kerbis Peterhans, Stanley, Webala, Gnoske, Hackett and Stanhope2016; Boundenga et al., Reference Boundenga, Ngoubangoye, Mombo, Tsoumbo, Renaud, Rougeron and Prugnolle2018) with the exceptions of the studies in South Sudan and Central Gabon (Schaer et al., Reference Schaer, Perkins, Ejotre, Vodzak, Matuschewski and Reeder2017; Rosskopf et al., Reference Rosskopf, Held, Gmeiner, Mordmüller, Matsiégui, Eckerle, Weber, Matuschewski and Schaer2019). The study in South Sudan revealed Hepatocystis infections in over 80% of the investigated epauletted fruit bats and in contrast, the study in Central Gabon reported no haemosporidian infections in the epauletted fruit bats (Schaer et al., Reference Schaer, Perkins, Ejotre, Vodzak, Matuschewski and Reeder2017; Rosskopf et al., Reference Rosskopf, Held, Gmeiner, Mordmüller, Matsiégui, Eckerle, Weber, Matuschewski and Schaer2019). For the first time, a Hepatocystis infection is reported for the pteropodid host R. aegyptiacus. Compared to the commonly infected epauletted fruit bats that roost in small groups in trees, R. aegyptiacus features a different roosting behaviour, forming large cave-dwelling colonies (e.g. ACR., 2017). To date, only Plasmodium infections have been documented for this bat species. Plasmodium (Vinckeia) rousetti was described from Rousettus leachi (old synonym for R. aegyptiacus) in the Democratic Republic of the Congo (van Riel and Hiernaux-L'Hoest, Reference van Riel and Hiernaux-L'Hoest1951) and subsequently reported from the same species (de Faveaux, Reference de Faveaux1958; Garnham, Reference Garnham1966). Apart from these three studies, which date back well over 50 years ago, no infections with haemosporidian parasites have been re-discovered in R. aegyptiacus (e.g. Schaer et al., Reference Schaer, Perkins, Decher, Leendertz, Fahr, Weber and Matuschewski2013; Lutz et al., Reference Lutz, Patterson, Kerbis Peterhans, Stanley, Webala, Gnoske, Hackett and Stanhope2016; Boundenga et al., Reference Boundenga, Ngoubangoye, Mombo, Tsoumbo, Renaud, Rougeron and Prugnolle2018; Rosskopf et al., Reference Rosskopf, Held, Gmeiner, Mordmüller, Matsiégui, Eckerle, Weber, Matuschewski and Schaer2019). Strikingly, here we detected Hepatocystis parasites, and not the previously reported Plasmodium species in this bat host. The close phylogenetic relationships of Plasmodium and Hepatocystis parasites (Galen et al., Reference Galen, Borner, Martinsen, Schaer, Austin, West and Perkins2018) raise the possibility that the previous reports of P. rousetti might instead be Hepatocystis infections, as reported herein. However, the morphological description of Plasmodium in R. aegyptiacus was based on the detection of schizont stages in the blood (van Riel and Hiernaux-L'Hoest, Reference van Riel and Hiernaux-L'Hoest1951), a life cycle stage that is lacking in Hepatocystis parasites. Additional investigations of R. aegyptiacus are needed to gain a better understanding of what appear to be rare infections of Hepatocystis parasites in this host species. The low prevalence of Hepatocystis in this study, and the lack of positive records in previous studies suggests that R. aegyptiacus might not represent a common host for Hepatocystis, and that detected cases are rare events that may perhaps be related to vector feeding preferences.
The blood smear of the infected R. aegyptiacus individual was of poorer quality than usual, as it was prepared after two days of blood storage. However, several mature gametocytes were readily detectable, and the morphology complies with descriptions of Hepatocystis (Fig. 2C–f, i). The presence of gametocytes in the blood confirms R. aegyptiacus as a true host as these life cycle stages present the proof that the parasite has completed its life cycle in the tissue and subsequently in the blood of the vertebrate host. Most importantly, the gene sequences unambiguously identify the infection as belonging to Hepatocystis. By sequencing different nuclear markers for the infected R. aegyptiacus we confirmed the morphological identification of the host sample as R. aegyptiacus.
Bats of the genus Rhinolophus as well as emballonurid bats have been reported as hosts of Nycteria parasites (Perkins and Schaer, Reference Perkins and Schaer2016). In this study, no haemosporidian parasites were detected in samples of the two insectivorous bat species C. afra and Rhinolophus sp. However, the sample size for these species was considerably small, a more elaborate cross-sectional sampling of the insectivorous bats in the area is needed to further investigate haemosporidian parasites in this bat group.
In conclusion, a survey of haemosporidian parasites in bats in two consecutive years in a protected savannah habitat in the Jos plateau, Nigeria, revealed abundant Hepatocystis infections in pteropodid hosts and uncovered a previously unknown competent host, R. aegyptiacus, for complete Hepatocystis blood stage maturation.
Supplementary material
The supplementary material for this article can be found at https://doi.org/10.1017/S0031182019000817
Author ORCIDs
Juliane Schaer, 0000-0001-6714-5771.
Acknowledgements
We would like to thank Tigga Kingston, Kendra Phelps, Iroro Tanshi, Chima Nwaogu and Joshua Kamani. We further want to thank John Azi for assistance in the field.
Financial support
This work was supported in part by the Max Planck Society and the Humboldt University Berlin.
Conflict of interest
None.
Ethical standards
The responsible institution for the approval of the use of Amurum Forest Reserve for research, the Scientific and Research Ethics Committee of the A. P. Leventis Ornithological Research Institute (APLORI), reviewed and officially approved this survey (4 May 2015).





