Introduction
Recently, a growing number of studies have been focusing on the interaction between nutrition and gene expression. Nutrients can interact with the genome directly (via nutrient response elements) and indirectly via modulation of mechanisms including DNA methylation, histone modification and non-coding RNA, in particular microRNA (miRNA). To date, DNA methylation and histone modifications are the best characterised of these mechanisms, with investigation into the links between nutrition and miRNA expression only being considered much more recently, and in much less depth.
Although both macro- and micronutrients can potentially modulate these mechanisms, targeting micronutrients is likely to be the most viable avenue for potential therapeutic intervention, as this avoids the confounding factor of energy provision by macronutrients. As such, the purpose of this review is to present the current scientific data suggesting that micronutrients can modulate the expression of miRNA. miRNA signatures and aberrant expression profiles have been linked to a range of disease states, including those associated with ageing, life-style and dietary factors( Reference Papageorgiou, Tousoulis and Androulakis 1 – Reference Calin and Croce 3 ). Therefore, modulation of miRNA profiles may explain an additional link between molecular nutrition and phenotypes of health and disease. While the investigation of the role of miRNA in disease is not novel, investigation of the interactions between micronutrients, miRNA and disease processes is a relatively nascent area of investigation, and much remains to be elucidated.
MicroRNA
miRNA are an abundant class of short (about eighteen to twenty-three nucleotides), non-coding RNA, which are involved in post-transcriptional regulation of gene expression. The first description of a miRNA was that of lin-4 in Caenorhabditis elegans by Lee et al. in 1993( Reference Lee, Feinbaum and Ambros 4 ). Following the identification of multiple similar small regulatory RNA in plants and animals, the term microRNA was coined( Reference Ruvkun 5 ). To date, over 2000 mature human miRNA have been identified (MiRBase, release 20, June 2013; http://www.mirbase.org); however, the specific function of the majority of these has not yet been ascertained.
Due to their small size, each miRNA may target hundreds of messenger RNA (mRNA)( Reference Selbach, Schwanhausser and Thierfelder 6 ). Likewise, each mRNA can be targeted by multiple miRNA, allowing complex and multifaceted regulatory networks( Reference Brennecke, Stark and Russell 7 ). It has been estimated that miRNA have a post-transcriptional regulatory influence on more than half of all human mRNA( Reference Friedman, Farh and Burge 8 ). As such, they are believed to participate in almost all cellular processes.
A majority of investigations have centred on the role of miRNA in cancers( Reference Calin and Croce 3 ); however, aberrant miRNA expression profiles have been identified in a range of conditions including asthma( Reference Ariel and Upadhyay 9 ), CVD( Reference Papageorgiou, Tousoulis and Androulakis 1 ), autoimmune disease( Reference Wang, Peng and Ouyang 10 ) and diabetes( Reference Lorenzen, Kumarswamy and Dangwal 2 ). Unique signatures have been identified in diseased tissues (relative to adjacent healthy tissue or healthy control subjects( Reference Calin and Croce 3 )) and, more recently, plasma and serum miRNA signatures have been connected to particular disease states (for a review, see Weiland et al. ( Reference Weiland, Gao and Zhou 11 )). These circulating miRNA are promising objective and non-invasive biomarkers for disease diagnosis and prognosis, and potentially even nutritional status. Circulating miRNA offer many potential advantages as biomarkers. Circulating miRNA can be detected in blood with high levels of specificity and sensitivity, and are remarkably stable. As such, they are practical biomarkers detecting or staging a wide range of pathological processes( Reference Gilad, Meiri and Yogev 12 ). However, additional research is needed to unravel the complex miRNA profiles associated with pathophysiological conditions before this promise is fully realised.
The biogenesis and complex action of miRNA have been comprehensively reviewed elsewhere( Reference Bartel 13 , Reference Patil, Zhou and Rana 14 ) and details are outside the scope of the present review. Briefly, miRNA (as part of the RNA-induced silencing complex; RISC) act on mRNA via complementary base pairing, usually to repress translation or to increase the degradation of the target mRNA (Fig. 1). In cases of near-complete miRNA–mRNA complementarity, the target mRNA may be directly cleaved by argonaute proteins( Reference Yekta, Shih and Bartel 15 , Reference Davis, Caiment and Tordoir 16 ). More commonly, partial complementarity results in modifications that repress translation or reduce the stability of the target mRNA( Reference Chendrimada, Finn and Ji 17 – Reference Wu, Fan and Belasco 19 ). Interestingly, more recent reports have demonstrated a capability of miRNA to activate target translation under specific conditions, such as during cell-cycle arrest( Reference Vasudevan, Tong and Steitz 20 – Reference Vasudevan 23 ).
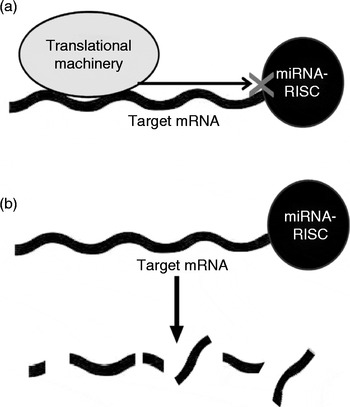
Fig. 1 MicroRNA (miRNA) mechanisms of action. miRNA can act by (a) blocking gene transcription or (b) inducing miRNA instability or degradation. mRNA, messenger RNA; RISC, RNA-induced silencing complex.
MicroRNA in epigenetics
The definition of epigenetics is often varied and is continuously evolving( Reference Weichenhan and Plass 24 ). It remains a matter of contention as to whether miRNA themselves are technically a component of the epigenome, or a complementary player. Most generally, epigenetics is defined as heritable changes in gene expression that do not involve a change in DNA sequence( Reference Chuang and Jones 25 – Reference Sato, Tsuchiya and Meltzer 27 ). Under this broad description, if gene expression is regarded as the generation of a finally functional protein product of a gene, and not merely transcription of a gene, it is possible to argue for the inclusion of miRNA in the definition of epigenetics. However, others specify that the consensus definition of an epigenetic trait is a phenotype resulting from changes to the chromosome, in the absence of any alteration to the DNA sequence( Reference Berger, Kouzarides and Shiekhattar 28 ). Under this definition it is impossible to include miRNA in the epigenome, as their gene-regulation potential occurs post-transcriptionally. In this context, miRNA have been described as epigenetic ‘initiators’( Reference Berger, Kouzarides and Shiekhattar 28 ).
Regardless of whether miRNA are positioned within or peripheral to the definition of epigenetics, the two are inextricably linked( Reference Weichenhan and Plass 24 ). miRNA and the epigenome can regulate each other, as well as act in concert, forming a complex bi-directional epigenetics–miRNA regulatory circuit. Importantly, miRNA and other epigenetic modifications are all plastic in response to stimuli, such as environmental exposures and endogenous signalling. This ultimately allows unique phenotypes to stem from one genotype( Reference Park, Friso and Choi 29 ) via modification of gene expression potential( Reference Jaenisch and Bird 30 ).
Common epigenetic modifications can be classified as DNA methylation and histone modifications. DNA methylation is the most extensively studied epigenetic trait. DNA methylation occurs when methyl groups are added to the DNA sequence, normally at a CpG dinucleotide. Methylation in the promoter or other regulatory regions of a gene is usually associated with gene silencing( Reference Deaton and Bird 31 ). Multiple histone modifications (including acetylation, phosphorylation, ubiquitination and methylation of histone protein tails) can occur that regulate the transcription level of genes( Reference Das and Tyler 32 ). Importantly, none of these processes occurs independently and it is the sum of all of these marks, referred to as the epigenome, that modulates gene expression.
These mechanisms are important in normal processes, especially those that occur during development and ageing, but can also drive pathophysiological processes. It is thought that the modification of epigenetic marks on the genome explains how environmental and life-style factors can modify an individual's risk of developing certain diseases( Reference Park, Friso and Choi 29 ). The plasticity of epigenetic marks has led to significant interest in manipulation of the epigenome for therapeutic purposes( Reference Costello, Krzywinski and Marra 33 , Reference Christensen, Houseman and Marsit 34 ). miRNA are deeply involved in the epigenome. Not only can they directly modulate gene expression post-transcriptionally, but several miRNA (known as epi-miRNA) have been identified that directly influence the function of the other components of the epigenetic machinery( Reference Fabbri and Calin 35 ). This, in turn, indirectly influences the epigenetic modulation of other genes (Fig. 2). Potential nutritional regulators have been identified for some of these epi-miRNA (Table 1). Conversely, miRNA expression can also be regulated by classical epigenetic mechanisms (methylation and histone modifications; for a review, see Sato et al. ( Reference Sato, Tsuchiya and Meltzer 27 )) and expression of one miRNA may also regulate expression of another miRNA( Reference Martello, Rosato and Ferrari 36 ). The interaction and overlapping of these two mechanisms allow for regulation of miRNA levels and thus their genetic targets via complex feedback loops.
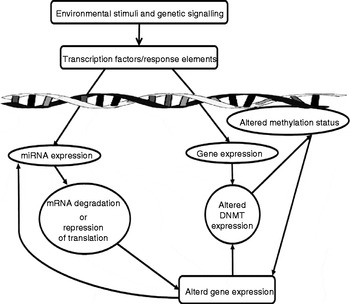
Fig. 2 Summary of the potential effects of vitamins and minerals on gene expression. MicroRNA (miRNA) are intrinsically involved in gene regulation. Vitamins and minerals can induce the expression of miRNA via the activation of transcription factors/response elements, leading to altered gene expression by the induction of messenger RNA (mRNA) degradation or the repression of translation. Vitamins and minerals may also alter the function of the classical epigenetic machinery, altering expression of DNA methyltransferases (DNMT) and histone modification enzymes, such as histone deacetylases and histone acetyltransferases. Modulation of these enzymes leads to changes in DNA methylation status and histone modifications, which in turn can modulate the expression of other genes, including miRNA themselves. These interconnecting pathways allow for the modulation of complex processes and feedback loops.
Table 1 Epigenetic microRNA with identified vitamin or mineral regulators
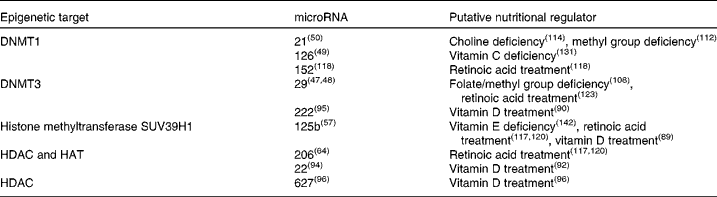
DNMT, DNA-methyl transferase; HDAC, histone deacetylase; HAT, histone acetyltransferase.
Several miRNA loci have been shown to be differentially methylated in a range of cancers( Reference Lujambio, Calin and Villanueva 37 – Reference Guan, Yin and Li 40 ). Examples of aberrant methylation of miRNA loci include miR-9-1 in breast cancer( Reference Lehmann, Hasemeier and Christgen 41 ) and miR-1 in hepatocellular carcinoma( Reference Datta, Kutay and Nasser 42 ). Treatment of the cancer cell line T24 with a DNA methylation inhibitor (5-aza-2′-deoxycytidine) resulted in increased expression of miR-126, -132 and -205( Reference Saito, Liang and Egger 39 ), suggesting that these miRNA were previously inhibited by methylation. Epigenetic drugs, targeting DNA methylation and histone modifications, are currently undergoing clinical trials and some have been approved by the Food and Drug Administration and the European Medicines Agency( Reference Nebbioso, Carafa and Benedetti 43 ). These interventions are likely to make an impact upon miRNA profiles and, as such, studies are needed to elucidate these treatment effects on miRNA profiles.
Treatment with histone deacetylase (HDAC) inhibitors, such as phenyl butyric acid, has also been shown to modulate miRNA profiles in multiple cancer cell lines( Reference Saito, Liang and Egger 39 , Reference Nasser, Datta and Nuovo 44 , Reference Scott, Mattie and Berger 45 ). This suggests that changes in gene expression previously attributed solely to the direct action of histone modifications may, at least in part, be due to the associated indirect effects of miRNA expression. miRNA may be involved in co-regulation or fine tuning of these processes or be a consequence of the histone modifications themselves. In particular, HDAC1 has been demonstrated to directly enhance miRNA expression by enhancing the rate of miRNA production via deacetylation of critical lysine residues in the RNA-binding domain of DGCR8 (DiGeorge syndrome critical region 8), a protein involved in miRNA biogenesis( Reference Wada, Kikuchi and Furukawa 46 ). This demonstrates that HDAC are involved in gene silencing both via transcriptional repression and by increasing miRNA abundance.
Multiple epi-miRNA have been identified that directly and indirectly target DNA-methyl transferases (DNMT), the enzymes responsible for DNA methylation( Reference Fabbri, Garzon and Cimmino 47 – Reference Duursma, Kedde and Schrier 51 ) as well as the enzymes responsible for histone modifications, such as histone methyltransferases( Reference Sander, Bullinger and Klapproth 52 – Reference Fan, Tsang and Tam 57 ), deacetylases( Reference Datta, Kutay and Nasser 42 , Reference Nasser, Datta and Nuovo 44 , Reference Sluijter, van Mil and van Vliet 58 – Reference Buurman, Gürlevik and Schäffer 63 ) and acetyltransferases( Reference Roccaro, Sacco and Jia 64 ). These studies, in cancer and inflammatory cells, have demonstrated that aberrant expression of multiple miRNA can alter the levels of DNMT and histone-modifying enzymes. This may have implications for the role of miRNA modulation in the pathogenesis and progression of cancers and inflammatory diseases.
It is likely that as investigations continue, more examples of epi-miRNA and epigenetically regulated miRNA will be identified. Despite the general acceptance of miRNA as important regulators of gene transcription, much remains to be determined about their genetic and environmental modulation and their consequences for health, disease and longevity. As several dietary factors are known to influence the epigenome, modulation of miRNA profiles via dietary stimuli warrants further discussion.
Nutritional modulation of epigenetic traits
Whilst the epigenome is plastic in response to stimuli, modifications may also persist long after the cessation of exposure. The role of epigenetics in underpinning the developmental origins of disease (the Barker hypothesis), which suggests that events occurring in utero can predispose to disease such as coronary artery disease and diabetes later in life( Reference Barker 65 – Reference Barker, Winter and Osmond 67 ), is now being recognised( Reference Canani, Costanzo and Leone 68 ). Differential methylation of disease-related genes have been identified in adults conceived during the Dutch famine of the Second World War, compared with their siblings conceived outside of famine( Reference Tobi, Slagboom and van Dongen 69 – Reference Heijmans, Tobi and Stein 71 ). Additional analyses of other epigenetic traits, including the role of miRNA in this population, are likely to be informative.
Studies of monozygotic twins have shown that differences in epigenetic traits appear to become more pronounced with ageing and divergence of life-style( Reference Fraga, Ballestar and Paz 72 – Reference Zhao, Goldberg and Vaccarino 75 ). As such, it is now realised that developmental and epigenetic plasticity extends beyond the pre- and postnatal periods and into later life. Ageing, genetics and environmental (including nutritional) influences all make an impact upon epigenetic regulation, with environmental influences the only practical point for intervention. As there are optimal nutritional requirements for maintaining the metabolome, and nutritional status has an impact upon physiology throughout life, nutrition clearly forms a significant environmental component( Reference Park, Friso and Choi 29 ).
Diet is therefore one of the most important modifiable determinants of risk in many diseases, with the phenotypic outcomes of adverse dietary habits accumulating over a lifetime. Chronic diseases or diseases with age-related onset, such as cancer, CVD and diabetes, are particularly well recognised as having associated life-style risk factors, in particular dietary components( Reference Roberts and Barnard 76 , 77 ). Diet also plays an additional ongoing role in disease, as patients suffering from chronic diseases may have impaired nutritional status as a consequence of disease( Reference Teunissen, Wesker and Kruitwagen 78 ). Roles for both dietary factors and miRNA have been identified in the functioning and modification of multiple disease-related pathways including DNA methylation, inflammation, cell-cycle repair, apoptosis and DNA repair. This has led to expanding investigations into the relationships between miRNA signatures, dietary habits and disease risk.
Modulation of microRNA expression by micronutrients
Manipulation of miRNA profiles through dietary modifications and supplements has been proposed as a potential future therapeutic intervention or prevention strategy( Reference Witwer 79 – Reference McKay and Mathers 81 ). While both macronutrients and micronutrients have been identified as potential disease and miRNA modifiers, micronutrients may represent the preferred intervention path, as this avoids the confounding role that macronutrients play in energy provision. Intervention, whether through dietary modifications or supplementation, may offer novel therapeutic strategies for a host of conditions. Despite the clear opportunities for novel preventative and therapeutic strategies, investigation into the dietary modulation of miRNA profiles remains in its infancy. To date, limited studies have been conducted in human cohorts, with the majority of studies conducted in cell systems or animal models. Details of known associations between miRNA and vitamins and minerals are summarised in online Supplementary Table S1 and described below.
Vitamin D
Vitamin D is a fat-soluble steroid vitamin and pro-hormone that can be consumed in the diet as cholecalciferol (vitamin D3) or ergocalciferol (vitamin D2), or synthesised by the skin in response to sunlight. In addition to its classical role in maintaining bone health and Ca homeostasis, epidemiological evidence has now identified a potential role for vitamin D deficiency in a range of conditions including diabetes( Reference Pittas, Lau and Hu 82 ), CVD( Reference Wang, Pencina and Booth 83 ), autoimmune disease( Reference Adorini 84 ) and some cancers( Reference Lappe, Travers-Gustafson and Davies 85 ). Vitamin D supplementation is now being investigated in the treatment and prevention of these diseases, with a particular focus on its potential as a cancer chemopreventative, due to the identified role of the vitamin D receptor (VDR) in cell-cycle regulation and differentiation( Reference Lamprecht and Lipkin 86 ).
The active vitamin D metabolite, calcitriol (1,25-dihydroxyvitamin D3), has long been known to directly regulate gene transcription via interaction with the VDR, which acts as a nuclear receptor transcription factor( Reference Carlberg 87 ). However, post-transcriptional regulatory mechanisms for vitamin D have also been proposed. Regulation of mRNA levels via miRNA signalling is now becoming recognised as a potential additional mechanism for action for vitamin D, and consequences of these interactions are now being investigated( Reference Sonkoly, Lovén and Xu 88 , Reference Giangreco, Vaishnav and Wagner 89 ).
Abnormal proliferation is a signature feature of cancer cells. Vitamin D has been demonstrated to suppress proliferation in multiple malignant cell lines( Reference Essa, Reichrath and Mahlknecht 90 – Reference Mohri, Nakajima and Takagi 93 ). In colon cancer cell lines (SW80-ADH and HCT116) the anti-proliferative effects of calcitriol treatment correlated with an induction of miR-22, -146a and -222 expression, and a reduction in miR-203 expression. miR-203 is a known suppressor of the proto-oncogene JUN ( Reference Sonkoly, Lovén and Xu 88 ). miR-222 and -22 are known epi-miRNA, targeting DNMT3 and HDAC4, respectively( Reference Zhang, Yang and Yang 94 , Reference Lee, Jeong and Lim 95 ). In particular, miR-22 expression was found to be induced in a dose-, time- and VDR-dependent manner. Moreover, miR-22 expression has also been shown to be lower in human colon cancer tissue, compared with surrounding normal tissue, correlating with changes in VDR expression levels( Reference Alvarez-Díaz, Valle and Ferrer-Mayorga 92 ). This suggests that miR-22 may have an anti-cancer effect that is modulated by vitamin D and the VDR. However, in another colorectal cancer cell line (HT-29 cells), miR-627 was the only miRNA significantly up-regulated by calcitriol treatment. Blocking miR-627 inhibited the anti-proliferative effects of calcitriol treatment. miR-627 targets Jumonji domain containing 1A (JMJD1A), which encodes a histone demethylase( Reference Padi, Zhang and Rustum 96 ).
Additional anti-cancer properties of vitamin D have been investigated in a range of cancer cell lines. Treatment of promyeloblastic leukaemia (HL60) and pro-monocytic leukaemia (U937) cells with low concentrations of calcitriol led to decreased expressions of miR-181a and -181b, in a dose- and time-dependent manner, and the arrest of cell-cycle progression in the G1 phase. Transfection of pre-miR-181a blunted these effects. Treatment of prostate cancer cell lines (RWPE-1, RWPE-2, PrEC and PrE) with calcitriol led to a significant up-regulation of the tumour-suppressor miRNA miR-100 and -125b( Reference Giangreco, Vaishnav and Wagner 89 ). In LNCaP prostate cancer cells, calcitriol treatment induced expression of miR-98, a tumour-suppressor miRNA, and suppressed proliferation. This occurred both via direct induction due to the enhanced binding of VDR to the vitamin D response element (VDRE) in the miR-98 promoter region, and indirectly via down-regulation of gene expression( Reference Ting, Messing and Yasmin-Karim 97 ). VDR–VDRE interaction has also been shown to up-regulate let-7a-2 expression in human lung cancer cells (A549 cells)( Reference Guan, Liu and Chen 98 ). Despite the diversity of the miRNA involved, these results suggest that aberrant expression of miRNA may contribute to the malignant phenotype and that this can be moderated by vitamin D treatment( Reference Wang, Gocek and Liu 99 ).
Vitamin D is also believed to play a role in protection from cellular stress. In a study using a breast epithelial cell line (MCF12F), calcitriol treatment protected cells against death in models of starvation, oxidative stress, hypoxia and apoptosis induction. Serum starvation led to significant increases in the expression of multiple miRNA (including miR-26b, -182, -200b/c, and the let-7 family). However, this was reversed in the presence of calcitriol, indicating that miRNA expression may be a mechanism that protects against cellular stress via a vitamin D-related process( Reference Peng, Vaishnav and Murillo 100 ). This may have implications in multiple disease states including diabetes, CVD and cancers( Reference Fulda, Gorman and Hori 101 ).
Although the evidence generated in cell studies suggests a role for miRNA and vitamin D in modulating disease-related processes, these studies often used supra-physiological doses. Furthermore, limited studies involving correlation between serum levels of vitamin D and miRNA expression in human subjects have been conducted. In a study of forty subjects given oral vitamin D supplementation for 12 months compared with thirty-seven receiving placebo, serum miRNA and vitamin D levels were measured at baseline and post-supplementation. At baseline a positive correlation was detected between serum 25-hydroxyvitamin D levels and miR-532-3p expression. miR-532-3p has been shown to target the gene for glucose-6-phosphatase, an important enzyme in glucose homeostasis( Reference Jia, Cong and Li 102 ). After 12 months of supplementation there was a significant difference in the expression of miR-221; however, this was due to a decrease in levels in the control, rather than the experimental, group( Reference Jorde, Svartberg and Joakimsen 103 ), possibly indicating that vitamin D deficiency in the placebo group led to the change in miR-221 expression.
In a study of pregnant women, plasma calcitriol was measured and patients were categorised as either low ( ≤ 25.5 ng/ml) or high ( ≥ 31.7 ng/ml)( Reference Enquobahrie, Williams and Qiu 104 ). Respectively, these groups reflect deficiency and sufficiency( Reference Nowson, McGrath and Ebeling 105 ). A total of eleven miRNA were differentially regulated (ten down and one up) in the low group compared with the high group. However, this study was of limited power, as only thirteen subjects were assayed and no functional conclusions were drawn( Reference Enquobahrie, Williams and Qiu 104 ).
Folate and related vitamins involved in methyl group metabolism
Folate (as the 5-methyltetrahydrofolate vitamer), methionine, choline and vitamin B12 are critical in one-carbon metabolism, which is the major source of methyl donors for cellular methylation reactions. These reactions include the essential processes of DNA synthesis and repair, DNA methylation, cell proliferation and the synthesis of amino acids( Reference Lucock 106 ). Folate can be used in the de novo generation of methionine and, as such, consumption of folate and other methyl group donors dictates the availability of methyl groups for use in methylation reactions. Therefore, folate and other methyl group donor levels may influence miRNA profiles indirectly via alteration of DNA methylation states, or may have an impact on miRNA expression levels through other methylation reactions. Low folate consumption has been associated with increased risks of birth defects, CVD, depression, hearing loss, and some cancers, particularly breast, colon, pancreatic and stomach( Reference Lucock 106 , Reference Lucock and Daskalakis 107 ).
Folate-deficient mouse embryonic stem cells have been shown to have inhibited growth, and a significantly higher rate of apoptosis. Multiple miRNA (let-7a, miR-15a, -15b, -16, -29a, -29b, -34a, -130b, -125a-5p, -124, -290 and -302a) were confirmed to be differentially expressed during folate deficiency( Reference Lianga, Lib and Liub 108 ). A similar magnitude of modulation has been demonstrated in human cell lines. Exposure of the human lymphoblast cell line TK-6 to folate-deficient media for 6 d led to a significant modulation of expression of twenty-four different miRNA. Importantly, return of these cells to folate-sufficient media led to a restoration of miRNA profiles to control levels( Reference Marsit, Eddy and Kelsey 109 ). Of particular interest in this study was the up-regulation of the epi-miRNA miR-222 and -22 under folate-deficient conditions. This in vitro result was complemented by the demonstration of higher levels of both these miRNA in human blood taken from patients in the lowest percentile for folate intake, compared with those in the highest percentile( Reference Marsit, Eddy and Kelsey 109 ). However, this study was conducted in a subset of patients from a larger case–control study of head and neck squamous cell carcinoma, which may be a confounding factor, and only included eleven subjects in total( Reference Marsit, Eddy and Kelsey 109 ).
Research on folate deficiency in animal models has centred on the role of methyl groups in hepatic cancer. Several animal models have demonstrated disease consequences of methyl group deficiency and have linked these to aberrant miRNA expression profiles. A methyl donor-deficient diet in rats leads to hepatocellular carcinoma (HCC) after 54 weeks( Reference Tryndyak, Ross and Beland 110 – Reference Kutay and Bai 112 ). Comparison of miRNA microarray profiles in liver tissue from rats fed this diet showed an increased expression of let-7a, miR-21, -23, -130, -190 and -17-92, and decreased miR-122, compared with rats fed the control diet( Reference Kutay and Bai 112 ). This adds weight to the changes in let-7a and miR-130b expression seen in cell-culture models. Interestingly, when the diet was returned to the control regimen at 36 weeks, miR-122 levels returned to normal and HCC did not develop( Reference Kutay and Bai 112 ).
Further analysis of this rat model identified profound down-regulation of miR-34a, -16a, -181a, -200b and -127, all of which are known tumour-suppressor miRNA. Notably, suppression of miR-34a and -127 occurred early and continued throughout carcinogenesis. This correlated with increased levels of E2F3 (E2F transcription factor 3) and BCL6 (B-cell lymphoma 6), proteins known to be regulated by miR-34a and − 127, respectively( Reference Tryndyak, Ross and Beland 110 , Reference Pogribny, Tryndyak and Ross 113 ).
In a murine model of a choline-deficient and amino acid-defined diet, hepatocarcinogenesis is usually induced after approximately 84 weeks( Reference Wang, Majumder and Nuovo 114 ). The oncogenic miR-155, -221, -222 and -21 were shown to be up-regulated in hepatic tissue in this model. This correlated with a significant reduction in the expression of the tumour-suppressor genes hepatic phosphatase and tensin homologue (PTEN) and CCAAT/enhancer binding protein β (C/EBPB), which are targets of miR-21 and -155, respectively. Furthermore, the expression of miR-155, as measured by in situ hybridisation and real-time RT-PCR, correlated with diet-induced histopathological changes in the liver( Reference Wang, Majumder and Nuovo 114 ).
Given the link between dietary methyl group donors and regulation of DNA methylation, and the miRNA listed above, further investigation into epigenetic regulation of miRNA and the impacts upon their targets is clearly warranted.
Retinoic acid
Retinoic acid, a metabolite of vitamin A (retinol), is important in growth and development. Like vitamin D, most of its known functions are mediated through binding to nuclear receptors and subsequent modification of gene transcription. Retinoic acid acts by binding to the retinoic acid receptor (RAR), forms a heterodimer with the retinoid X receptor (RXR) and binds to DNA in regions known as retinoic acid response elements. Binding of the retinoic acid ligand to RAR alters its conformation, affecting the binding of other proteins that either induce or repress transcription of nearby genes – including the developmentally significant transcription factors coded for by Hox genes and several other target genes( Reference Duester 115 ). However, modulation of miRNA profiles is now under investigation as an additional mechanism of action.
Retinoic acid is known to modulate neural differentiation and is used in neuroblastoma therapies, where a potential role for miRNA has been identified. In NT-2 (human embryonic carcinoma) cells, differentiation into neural cells is induced by treatment with retinoic acid. However, when miR-23 expression is knocked down with a small interfering RNA, this process does not occur, indicating a mechanistic role for miR-23 in the differentiation process( Reference Kawasaki and Taira 116 ). Treatment of neuroblastoma cell lines with retinoic acid leads to cellular differentiation and inhibits proliferation( Reference Annibali, Gioia and Savino 117 – Reference Beveridge, Tooney and Carroll 119 ). miR-9 and -103a are both up-regulated in these cells following treatment. Both of these miRNA target ID2, a transcription factor expressed in neural precursor cells, but also up-regulated during tumorigenesis in the neural system( Reference Annibali, Gioia and Savino 117 ). Conversely, retinoic acid has also been shown to down-regulate miR-9, -124a and -125b expression in spinal tissue in a rodent model of spina bifida, suggesting that these miRNA may be involved in the modulation of embryonic spinal development( Reference Zhao, Sun and Wang 120 ). This may demonstrate differential expression in different tissues and environments, and highlights the need for further study.
Retinoic acid treatment of neuroblastoma cell lines has also been shown to lead to an increased expression of miR-152, which has been shown to target DNMT1. Over-expression of miR-152 in SK-N-BE cells mimicked some, but not all, of the features of differentiation seen with retinoic acid treatment, suggesting that miR-152 is only partially responsible for phenotype in this system( Reference Das, Foley and Bryan 118 ). The miR-17 family has been identified as down-regulated in retinoic acid-induced differentiation of neuroblasts( Reference Beveridge, Tooney and Carroll 119 ). miR-7 has been shown to be a regulator of both proliferative and anti-proliferative genes, possibly via the regulation of the mitogen-activated protein kinase (MAPK) signalling pathway( Reference Beveridge, Tooney and Carroll 119 , Reference Cloonan, Brown and Steptoe 121 , Reference Yang, Yin and Wang 122 ). This demonstrates a potential role for miR-17 in conjunction with other miRNA in the complex control of neuronal differentiation.
Retinoic acid induces granulocytic differentiation in leukaemia cell lines. In HL-60 cells, treatment with all-trans-retinoic acid (ATRA) modulates the expression of multiple miRNA, including the up-regulation of miR-29a, -142-3p( Reference Wang, Gong and Yu 123 ), -663, -494, -145, -22, -363* and -223( Reference Jian, Li and Fang 124 ). The findings for miR-29a and -142-3p were replicated in THP-1 and NB4 cells. Additionally, forced expression of either miRNA in haematopoietic stem cells from healthy controls and acute myeloid leukaemia (AML) patients promoted myeloid differentiation. This suggests that miR-29a and -142-3p are key regulators of normal myeloid differentiation and their reduced expression is involved in AML development( Reference Wang, Gong and Yu 123 ).
Treatment of NB4 and primary cells from acute promyelocytic leukaemia patients with ATRA up-regulated the expression of let-7d/a-3, miR-223, -107, -15a and -16-1, which is also negatively correlated with Bcl-2 (B-cell lymphoma 2) and Ras protein expression( Reference Garzon, Pichiorri and Palumbo 125 ). miR-16-1 and let-7a transfection demonstrated that these miRNA targeted the genes BCL-2 and RAS, respectively. Bcl-2 is a regulator of apoptosis and Ras regulates a range of cellular processes. Furthermore, the recruitment of NF-κB on the let-7a-3/let-7b cluster promoter upon treatment with retinoic acid suggests that this is the mechanism of regulation of miRNA expression in this case( Reference Garzon, Pichiorri and Palumbo 125 ). Conversely, authors of a later study of ATRA differentiation in NB4 cells reported that let-7a was down-regulated following ATRA treatment( Reference Rossi, D'Urso and Gatto 126 ). This apparent contradiction is probably due to the differences in culture time used in the two experiments (48 h( Reference Garzon, Pichiorri and Palumbo 125 ), compared with 6 d( Reference Rossi, D'Urso and Gatto 126 )). This highlights the importance of the temporal specificity of the relationships between nutrients, miRNA expression and physiological consequences and this needs to be considered in furthering these studies.
Vitamin C
Vitamin C (ascorbate, dehydroascorbate and monodehydroascorbate) is a cofactor in multiple biological processes, including collagen synthesis, neurotransmitter synthesis and hormonal activation. Vitamin C deficiency is perhaps best known for causing scurvy. Vitamin C is also an antioxidant molecule and as such it has been suggested to have a positive impact on CVD, hypertension, chronic inflammatory disease and diabetes. However, evidence for its health benefits is still inconclusive and further long-term studies are required( Reference Juraschek, Guallar and Appel 127 – Reference Riccioni, Frigiola and Pasquale 129 ).
Limited investigations have occurred into the influence of vitamin C on miRNA profiles, and none yet has targeted the disease states listed above. miRNA profiles in response to vitamin C status have, however, been investigated in hormonal regulation and cell differentiation. In periodontal ligament cells (a multi-potent cell population) treatment with ascorbic acid induced cellular differentiation and an increase in miR-146 expression. Stimulation of these cells with miR-146 alone led to the induction of differentiation( Reference Hung, Chen and Kuang 130 ). In mice deficient in the enzyme l-gulono-γ-lactone oxidase (unlike humans, mice can normally synthesise ascorbic acid in a pathway dependent on this enzyme) cells have reduced capacity to oppose oxidative stress( Reference Kim, Ku and Rosenwaks 131 ). In murine ovarian follicular cells, this corresponds with higher levels of let-7b, miR-16, -30a, -126, -143, -322 and -721 and lower levels of let-7c. The cause and consequence of these changes are yet to be identified, but suggest a potential role for vitamin C in regulating the target genes of these miRNA in developmental and maturation processes( Reference Kim, Ku and Rosenwaks 131 ). As the role of antioxidants such as vitamin C in chronic disease is further investigated, there is potential for additional roles for vitamin C in modulation of miRNA expression to be identified.
Vitamin E
Vitamin E belongs to a fat-soluble group of vitamins that includes several vitamers belonging to the tocopherols and tocotrienols. Vitamin E has multiple biological functions, including its role as a major lipid-soluble antioxidant, and, unlike many other vitamins, it is not an enzyme cofactor. A major role for vitamin E has also been identified in the regulation of gene expression in some instances. An inverse association between l-α-tocopherol (the major biologically active vitamer of vitamin E) intake and some cancers has been reported in multiple studies( Reference Woodson, Tangrea and Barrett 132 – Reference Zhang, Shu and Li 137 ). However, not all studies have drawn the same conclusions, with others finding a neutral( Reference Ocké, Bueno-de-Mesquita and Feskens 138 , Reference Gilbert, Metcalfe and Fraser 139 ) or detrimental effect on cancer and heart disease risk( Reference Lonn, Bosch and Yusuf 140 , Reference Miller, Pastor-Barriuso and Dalal 141 ). This lack of consensus may be due to a significant variance in the doses tested – a 2005 meta-analysis found that studies used a range of doses from 16.5 to 2000 IU/d( Reference Miller, Pastor-Barriuso and Dalal 141 ).
The impact of dietary α-tocopherol on miRNA expression has been investigated in a rodent model of vitamin E deficiency. miR-122 and -125b were selected for assessment due to their known roles in lipid metabolism, cancer progression and inflammation. Vitamin E deficiency resulted in decreased levels of both miR-122 and -125b, as well as reduced plasma cholesterol levels( Reference Rimbach, Moehring and Huebbe 142 ). Synthetic inhibition of miR-122 in mice led to the increased expression of 108 genes related to lipid metabolism and led to a significant reduction of plasma cholesterol( Reference Esau, Davis and Murray 143 , Reference Elmén, Lindow and Silahtaroglu 144 ). Although the effects of vitamin E depletion are not as strong as targeted inhibition, these studies suggest that vitamin E regulates gene expression at the post-transcriptional level through miRNA modulation; however, the gene and transcription factor targets are yet to be validated.
Selenium
Se is an essential nutrient found in Brazil nuts, shellfish, chicken, organ meats, game meat and beef. It exists in a range of chemical forms, including: selenomethionine, selenocysteine, selenate, selenoneine and selenite. Se has structural and enzymic biological roles, pro-apoptotic and DNA repair properties and is an important cofactor in the production and activity of antioxidant enzymes. Se is vital for human health and deficiencies have been linked to various diseases including cancers, autoimmune disease and CVD( Reference Rayman 145 ).
Association studies have suggested links between low Se status, cognitive decline, immune disorders and increased mortality. High Se levels appear to be beneficial in some cancer types, but the results of supplementation intervention trials have been mixed, suggesting that supplementation is only beneficial if dietary intake is inadequate( Reference Rayman 145 ). Some studies have suggested a positive correlation between Se status and risk of type 2 diabetes; however, other studies have shown no correlation( Reference Rayman, Blundell-Pound and Pastor-Barriuso 146 – Reference Laclaustra, Navas-Acien and Stranges 148 ).
miRNA modulation is a potential mechanism of action for Se; however, it is clear that additional empirical evidence is required to elucidate the potential of Se as a nutraceutical. Treatment of LNCaP human prostate cancer cells with selenite, a natural form of Se, has been shown to induce an up-regulation of p53 expression which occurs in parallel with an up-regulation of miR-34b/c( Reference Sarveswaran, Liroff and Zhou 149 ). The miR-34 is family known to target the p53 gene( Reference Hermeking 150 ) and this finding suggests that this miRNA family plays a role in modulation of the cell cycle in the induction of cancer. Despite this, further investigation is needed to determine the target of Se-induced miR-34 and any additional miRNA that may be influenced by Se status.
Zinc
Zn is required for numerous structural, catalytic and regulatory functions. Deficiency has been linked to growth retardation, dermatitis, taste impairment, delayed puberty, haematological abnormalities and immune dysfunction( Reference Prasad 151 ). In a study using varied dietary intakes of Zn (acclimatisation, depletion and repletion) in healthy adult males, nine miRNA were identified in plasma that were sensitive to plasma Zn concentrations. miR-10b, -155, -200b, -296-5p, -373, -92a, -145, -204 and -211 were all reduced in abundance during the depletion phase of the diet and increased during the repletion phase. Depletion corresponded to a compromised ability of blood cells to produce the inflammatory cytokine TNFα in response to inflammatory stimuli( Reference Ryu, Langkamp-Henken and Chang 152 ). The consequences of the modulation of the miRNA profile may in fact be more extensive and require further investigation.
Iron, magnesium and aluminium
Treatment of microglial cells with Fe and aluminium sulfate, aluminium sulfate alone, but not Fe sulfate alone has been shown to up-regulate miR-125b and -146a expression, via an NF-κB-dependent mechanism. These miRNA are involved in astroglial cell proliferation and inflammatory responses, respectively, and are significantly up-regulated in Alzheimer's disease( Reference Pogue, Percy and Cui 153 ). Modulation of specific miRNA by Fe alone has yet to be demonstrated; however, an important role for ferric haem has been demonstrated in the modulation of the miRNA processing machinery. DGCR8 forms a highly stable and active complex with ferric haem. When reduced to the ferrous state, the primary transcript (pri)-miRNA-processing capacity of the DGCR8 complex is abrogated( Reference Barr, Smith and Chen 154 ).
Likewise, Mg has been shown to interact with the miRNA-processing machinery, with RISC (RNA-induced silencing complex) being an Mg2+-dependent protein. Mg ions are also located at the small RNA-binding domain of the argonaute protein and are important for sequence-specific miRNA–target interactions, contributing to the binding of miRNA to the argonaute protein and thus playing a role in the cleavage of miRNA targets. Mg ions have also been shown to modulate the global stabilisation of the argonaute protein( Reference Ma, Xue and Zhang 155 ).
Direct uptake of microRNA from foods
It has recently been suggested that diet may have an even more direct impact upon miRNA profiles. The plant and animal foods included in our diet also contain miRNA that may be taken up into the body upon consumption, and as such may remain biologically active. If this is possible, then sequence homology could potentially allow regulation of endogenous genes by miRNA acquired from dietary sources( Reference Zhang, Hou and Chen 156 ). Previously, this cross-taxonomic regulation by small RNA had been demonstrated in a variety of model systems, including bacteria, viruses and worms( Reference Newmark, Reddien and Cebrià 157 – Reference Omarov and Scholthof 159 ). miRNA are also secreted in human breast milk and cows' milk, and may serve as a route of biological communication through feeding( Reference Zhou, Li and Wang 160 – Reference Kosaka, Izumi and Sekine 162 ).
Exogenous miRNA may originate from multiple sources. Plant miRNA remain detectable, although some at reduced levels, in cooked wheat, rice and potatoes( Reference Zhang, Hou and Chen 156 ). It has also been reported that the miRNA in cows' milk are stable under conditions that would normally be considered degenerative for miRNA( Reference Izumi, Kosaka and Shimizu 161 ). In future studies this needs to be verified for other foods and cooking conditions (methods, temperatures and times). It is hypothesised that other components of food, such as cholesterol and protein, may protect miRNA from degradation( Reference Jiang, Sang and Hong 163 ).
The first suggestion of the uptake of exogenous miRNA from food in humans has only recently been published. In 2012, Zhang et al. ( Reference Zhang, Hou and Chen 156 ) described in a study ten pools of serum from thirty healthy Chinese adults that almost thirty plant miRNA were detectable in serum, albeit at much lower levels than endogenous miRNA. Two of these, miR-156a and -168, are detectable in high levels in rice and crucifers, which are staple foods in the Chinese diet. Additionally, mice fed rice were reported to have elevated serum and tissue levels of miR-156a and -168, compared with those fed a standard mouse chow diet after 6 h. Furthermore, oral administration of mice with miR-168a isolated from fresh rice, synthetic miR-168a, or methylated synthetic miR-168 all led to increases in miR-168a levels in the serum and liver 3 h post-oral administration( Reference Zhang, Hou and Chen 156 ).
However, these findings are yet to be independently replicated, and others have since presented conflicting studies. It has now been suggested that there is in fact limited evidence for the general dietary uptake of exogenous miRNA from plants and questions have been raised regarding this hypothesis( Reference Witwer, McAlexander and Queen 164 , Reference Snow, Hale and Isaacs 165 ). After feeding a plant miRNA-rich substance to two pigtailed macaques, plant miRNA were only detected in plasma in some assays, at very low, and possibly non-specific levels and did not shift substantially from baseline following consumption of the plant miRNA-rich substance( Reference Witwer, McAlexander and Queen 164 ). Additional feeding studies high in exogenous miRNA in human subjects, mice and bees were also unable to detect significant levels of endogenous miRNA( Reference Snow, Hale and Isaacs 165 ). A study of publicly available small RNA datasets concluded that while plant RNA was identified in animal samples, this detection was at a low level and probably due to a sequencing artifact or external contamination( Reference Zhang, Wiggins and Lawrence 166 ). These studies suggest that the dietary uptake of miRNA is at best limited and may not be robust.
Furthermore, it has been suggested that the quantities of exogenous miRNA that would need to be ingested to elicit a systemic biological effect would need to be greater than is possible by dietary consumption( Reference Petrick, Brower-Toland and Jackson 167 ). Additional studies, perhaps utilising larger sample sizes and longer-term administration, and additional scrutiny of the methodologies employed, are needed to address the current discrepancies in the available data. It is yet to be definitively established whether or not exogenous miRNA can be taken up in sufficient levels to have consequences for gene expression in real-life situations. However, if this were possible, it would have significant implications for the analysis of potential dietary modulation of miRNA profiles, as past studies may have inadvertently identified exogenous miRNA as endogenous miRNA.
Conclusion
Investigation of the role of nutrition and diet in the modulation of miRNA profiles remains a relatively new field of investigation. Several observational studies and model systems have identified potential and logical links between nutritional status, miRNA profile and disease status; however, these links are, as yet, far from conclusive. In particular, additional studies in human cohorts are required.
Furthermore, the implications of identified associations cannot be fully appreciated without additional research to identify the mRNA targets of more miRNA, as the majority remain poorly defined. Additional functional studies are required in future research to understand if there are physiological consequences for modulation of miRNA by micronutrients and if these relationships can be harnessed for therapeutic or diagnostic purposes. The increasing affordability and practicality of miRNA assays are likely to contribute to an increased volume of investigation in coming years.
The proposal that plant miRNA can reach human circulation introduces an additional area for consideration, as, if proven, this may confound the interpretation of responses that are attributed to endogenous nutrition-related miRNA and may, in fact, contribute to pathophysiology. The identified roles for nutritional and dietary components suggest that dietary influences should be considered in future cohort studies attempting to link miRNA expression profiles with disease states. Micronutrients also offer a potential novel avenue for the development of therapeutic supplements that target miRNA modulation.
Supplementary material
To view supplementary material for this article, please visit http://dx.doi.org/10.1017/S0954422414000043
Acknowledgements
The present review was not funded by any granting body.
E. L. B. wrote the manuscript and was involved in its conceptualisation; Z. Y, M. V., K. D. and M. L. oversaw conceptualisation and implementation of the manuscript. All authors reviewed and approved the manuscript.
The authors do not have any conflict of interest.





