Article contents
Extensions to the model of an infinite number of selectively neutral alleles in a finite population
Published online by Cambridge University Press: 14 April 2009
Summary
The model, suggested by Ohta & Kimura, of an infinite number of selectively neutral alleles, in which alleles can only mutate to neighbouring alleles, has been extended to include a migratory pattern. The stepping-stone and island models of migration have been considered. In the latter case, it has been found that as the number of colonies becomes large, the expected number of alleles, ne, becomes approximately equal to
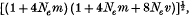
where m is the migration rate, v is the mutation rate and Ne is the effective size of each population. This solution tends to that of Ohta & Kimura as m → 0 but one can see that for any appreciable value of Nem, a large increase in ne is obtained. In order to check the validity of iterative results, models with a finite number of alleles have been considered, and their solutions have been found to converge quickly to those of the infinite case. The results exemplify the great power of migratory structure and neutral alleles to maintain a large amount of heterozygosity. Double step mutation and the finite time solution have also been considered.
- Type
- Research Article
- Information
- Copyright
- Copyright © Cambridge University Press 1975
References
REFERENCES
- 6
- Cited by


