One of the most important livestock animals in the food industry is the pig( Reference Dodson, Hausman and Guan 1 ). To reduce carcass fatness and improve feed efficiency, pigs reared in intensive systems have become leaner exhibiting fast growth. Leaner pigs display low intramuscular fat (IMF) content, a key meat quality trait, and sensory properties of pork are negatively affected when IMF is reduced below 2–2·5 %( Reference DeVol, McKeith and Bechtel 2 ).
Recent studies have used reduced protein diets (RPD) or low lysine levels as the most successful nutritional strategies to enhance fat accumulation in skeletal muscle without increasing subcutaneous adipose tissue in pigs( Reference Doran, Moule and Teye 3 – Reference Madeira, Pires and Alfaia 5 ). This finding relies on tissue-specific stimulation of lipogenic enzyme expressions under RPD, which in turn can lead to the increase of de novo fatty acid synthesis( Reference Doran, Moule and Teye 3 ). In addition, it is well known that dietary lysine deficiency reduces protein synthesis and increases the energy available for fat deposition( Reference Witte, Ellis and Mckeith 6 ). This mechanism can be related to higher transcription levels of PPARγ and sterol regulatory element-binding factor 1 (SREBF1) found in the muscle of growing pigs fed low dietary lysine, thus promoting lipogenesis( Reference Horton, Golstein and Brown 7 , Reference Schadinger, Bucher and Schreiber 8 ). However, the effect of RPD and low lysine levels on hepatic fatty acid metabolism remains to be elucidated.
Liver, together with skeletal muscle and adipose tissue, plays a key role in the regulation of lipid metabolism in pigs( Reference Corominas, Ramayo-Caldas and Puig-Oliveras 9 ). It is the principal site of cholesterol synthesis and fatty acid oxidation, whereas de novo lipogenesis occurs essentially in both liver and adipose tissue( Reference Nafikov and Beitz 10 ). Furthermore, the uptake of plasma NEFA released by adipose tissue is the predominant route by which fatty acids are supplied to the liver. Thus, plasma fatty acids should influence hepatic fatty acid composition and the expression of several proteins responsible for lipid metabolism( Reference Vallim and Salter 11 ).
Acetyl-CoA carboxylase α (ACACA), fatty acid synthase (FASN) and stearoyl-CoA desaturase (SCD or delta9 desaturase) are key lipogenic enzymes for fatty acid biosynthesis. Attending to the paramount role of liver in fatty acid elongation and desaturation, fatty acid desaturase 1 (FADS1, encoding for delta5 desaturase) and fatty acid desaturase 2 (FADS2, encoding for delta6 desaturase) are membrane-bound enzymes that catalyse the synthesis of PUFA( Reference Nakamura and Nara 12 ). Moreover, carnitine O-acetyltransferase (CRAT) is the rate-limiting enzyme of lipid catabolism, transporting fatty acid esters to the mitochondria for β-oxidation. The transcriptional regulators, SREBF, carbohydrate response element-binding protein (ChREBP), CCAAT/enhancer-binding protein α and PPAR appear to be the main enzymes responsible for hepatic fatty acid synthesis and degradation, and subsequent TAG production( Reference Zhao, Wang and Song 13 ). In addition, glucose and insulin play a critical role in transcriptional and post-transcriptional regulation of lipogenesis, through the activation of hepatic ChREBP and SREBF1c genes, respectively( Reference Denechaud, Bossard and Lobaccaro 14 ). The fatty acid binding protein 4 (FABP4) is accountable for fatty acid transport in adipocytes( Reference Hocquette, Gondret and Baéza 15 ).
We have previously shown that the molecular mechanisms regulating fat deposition in skeletal muscle and subcutaneous adipose tissue in pigs are tissue specific and genotype specific, and linked to the differential expression of genes encoding for lipogenic enzymes and transcription factors( Reference Madeira, Pires and Alfaia 5 ). In the present study, we question the effect of RPD under different genotypes (commercial cross-bred lean pigs v. autochthonous fatty pigs) on hepatic lipid metabolism, through the assessment of fatty acid profile and gene expression levels of key lipogenic enzymes and associated transcription factors.
Methods
Animals and experimental diets
This trial was conducted at the facilities of Unidade de Investigação em Produção Animal (Instituto de Investigação Agrária e Veterinária, UIPA-INIAV). All the experimental procedures involving animals were reviewed by the Ethics Commission of the CIISA/FMV and approved by the Animal Care Committee of the National Veterinary Authority (Direção-Geral de Alimentação e Veterinária, Portugal), following the appropriate European Union guidelines (2010/63/EU Directive). In all, twenty commercial cross-bred pigs (50 % Large White, 25 % Landrace and 25 % Pietrain) and twenty Alentejana purebred pigs, all entire males, were selected, with an average initial body weight of 60 (sd 2) kg. The Alentejano pig is a Portuguese autochthonous breed reared in the southern region of Portugal and is genetically similar to the Spanish Iberian breed with low capacity for lean tissue deposition( Reference Garcia-Vaverde, Barea and Lara 16 ). Moreover, this breed is characterised for having slow growth and high feed conversion ratio as well as high fat deposition. At present, there is a growing interest in high-quality products from the Alentejana pig from both producers and consumers( Reference Ramalho 17 ). Pigs were fed a standard concentrate diet from weaning until the beginning of the experiment. Animals were housed in two pens of four pigs each and one pen of two pigs per treatment (n 10). Pigs were fed individually twice a day and had free access to water. Throughout the experiment, pigs were weighted weekly just before feeding. Pigs from each genotype were randomly assigned to one of two diets in a 2×2 factorial arrangement (two genotypes and two dietary regimens).
Diets were isoenergetically formulated (16 MJ metabolisable energy/kg) and differed in crude protein and lysine contents: 17·5 % of crude protein and 0·7 % of lysine (normal protein diet, NPD) and 13·1 % of crude protein and 0·4% of lysine (RPD not equilibrated for lysine, RPD). Diets were analysed for DM by drying samples (n 4) at 100°C to a constant weight. N content was determined by the Kjeldahl method( 18 ) and crude protein was calculated as 6·25×N. Crude fibre was determined by the procedure described by the Association of Official Analytical Chemists (AOAC)( 18 ). Samples were extracted with petroleum diethyl ether, using an automatic Soxhlet extractor (Gerhardt Analytical Systems) to determine crude fat. Determination of ash and starch contents was carried out according to the procedures described in AOAC( 18 ) and in the study by Clegg( Reference Clegg 19 ), respectively. Gross energy in the feed was determined by adiabatic bomb calorimetry (Parr 1261; Parr Instrument Company). Fatty acid methyl esters (FAME) of feed samples were analysed by one-step extraction and transesterification, using heptadecaenoic acid (17 : 0) as the internal standard( Reference Sukhija and Palmquist 20 ). Total amino acids were extracted according to the method described by AOAC( 21 ). The extract was analysed by HPLC (Agilent 1100; Agilent Technologies) to quantify amino acids in the feed, including lysine, according to the procedure reported by Henderson et al. ( Reference Henderson, Ricker and Bidlingmeyer 22 ). The ingredients and detailed proximate and fatty acid composition of the diets are shown in Table 1.
Table 1 Diet formulation and chemical composition of experimental diets (n 4)Footnote *
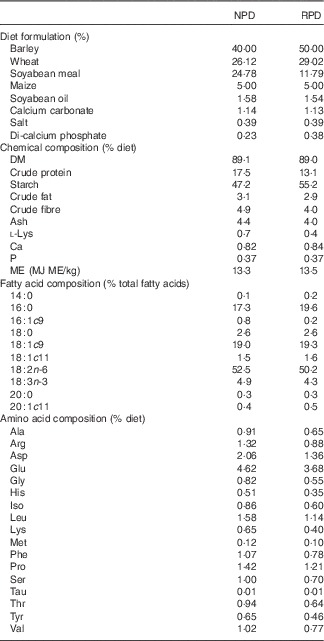
NPD, normal protein diet; RPD, reduced protein diet; ME, metabolisable energy.
* Others ingredients in both diets: pigs vitatec (0·4 %), tecaphos 500 g (0·1 %), ultracid V Dry EU (0·1 %), grain Tec TS (0·1 %), unilike Plus Dry (0·05 %), oxi-Nil Dry Premix (0·003 %).
Slaughter and sampling
Pigs were slaughtered at an average body weight of 93·4 (sd 2·42) kg at the INIAV experimental abattoir, after 17–19 h of fasting. After electrical stunning and exsanguination, blood samples were collected from the jugular vein and centrifuged at 1500 g for 15 min to obtain plasma. Samples for gene expression analysis were collected from the middle lobe of the liver, rinsed with sterile RNAse-free cold saline solution, cut into small pieces (thickness of approximately 0·3 cm), stabilised in RNA Later® solution (Qiagen) and stored at −80°C. For fatty acid composition, liver samples were vacuum-packed and stored at −20°C until analysis.
Plasma metabolites
Total cholesterol, HDL-cholesterol, LDL-cholesterol, TAG, phospholipids, total proteins, urea, N and glucose concentrations, aspartate aminotransferase, alanine aminotransferase (ALT), γ-glutamyltransferase (GGT) and alkaline phosphatase (ALP) were analysed using diagnostic kits (Roche Diagnostics) and a Modular Hitachi Analytical System (Roche Diagnostics). VLDL-cholesterol and total lipids were calculated as described by Friedewald et al. ( Reference Friedewald, Levy and Fredrickson 23 ) and Covaci et al. ( Reference Covaci, Voorspoels and Thomsen 24 ), respectively. Free fatty acids (FFA) were quantified using the Free Fatty Acid Quantification Kit (BioVision Inc.). Insulin and leptin concentrations were determined through the Porcine Insulin RIA kit (PI-12K; Linco Research) and the Multi-Species Leptin RIA kit (XL-85K; Linco Research), respectively. The degree of insulin resistance was calculated by the homoeostasis model assessment using the insulin resistance index (HOMA-IR): fasting serum glucose (mmol/l) times fasting serum insulin (mU/l) divided by 22·5( Reference Matthews, Hosker and Rudenski 25 ). Low HOMA-IR values indicate high insulin sensitivity, whereas high HOMA-IR values indicate high insulin resistance.
Hepatic lipid extraction and fatty acid composition
Liver samples were lyophilised (−60°C and 2·0 hPa), maintained exsiccated at room temperature and analysed within 2 weeks. Total lipids were extracted in duplicate and gravimetrically measured by the method described by Folch et al. ( Reference Folch, Lees and Stanley 26 ), using dichloromethane–methanol (2:1, v/v) instead of chloroform–methanol (2:1, v/v), as described by Carlson( Reference Carlson 27 ). Fatty acids were converted to methyl esters (FAME) by combined transesterification procedure with NaOH in anhydrous methanol (0·5 m), followed by HCl–methanol (1:1, v/v), at 50°C during 30 and 10 min, respectively, according to Raes et al. ( Reference Raes, De Smet and Demeyer 28 ). FAME were determined using a GC HP6890A (Hewlett-Packard), equipped with a flame ionisation detector and a CP-Sil 88 capillary column (100 m×0·25 mm i.d., 0·20-μm film thickness; Chrompack, Varian Inc.) using the conditions described in Alves & Bessa( Reference Alves and Bessa 29 ). The quantification of total FAME was carried out using nonadecanoic acid methyl ester (19 : 0) as the internal standard and by the conversion of relative peak areas into weight percentages. Fatty acids were identified on the basis of their retention times, corresponding to their FAME standards from Supelco Inc. and expressed as g/100 g of total fatty acids.
Hepatic RNA isolation and complementary DNA synthesis
Total RNA was isolated and purified from the liver using a modified protocol combining Trizol (Invitrogen) and RNeasy mini kit (Qiagen), respectively. Before real-time quantitative PCR (RT-qPCR), total RNA samples were treated with DNAse I (Qiagen). All the procedures were performed in accordance with the manufacturer’s protocols. RNA was quantified using a NanoDrop ND-2000c spectrophotometer (Nanodrop; Thermo Fisher Scientific). The A260/280 ratios were between 1·9 and 2·1. Reverse transcription was performed with a High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems). In brief, each 20 µl RT reaction contained 1 μg of DNase-treated total RNA template, 50 nm random RT Primer, 1×RT buffer, 0·25 mm of each dNTP, 3·33 U/µl multiscribe RT and 0·25 U/µl RNase inhibitor, and was subjected to heating at 25°C for 10 min, 37°C for 120 min and 85°C for 5 min. The complementary DNA (cDNA) solution obtained was divided into aliquots and stored at −20°C until further analysis.
Real-time quantitative PCR
Gene-specific intron-spanning primers were designed using Primer3 (http://frodo/wi.mit.edu/primer3/) and Primer Express Software version 2.0 (Applied Biosystems) based on Sus scrofa sequences (www.ncbi.nlm.nih.gov). Primers were purchased from NZYTech. Sequence homology searches against the database of GenBank showed that these primers matched only the sequence to which they were designed. To ensure optimal DNA polymerisation efficiency, the amplicon length ranged between 71 and 138 bp. Before performing qPCR assays, a conventional PCR was carried out for all genes in order to test the primers and verify the amplified products. To confirm identity of amplified fragments, PCR products were sequenced and homology searches were performed with Blast (www.ncbi.nlm.nih.gov/blast). In order to find the most stable endogenous control in the liver, five commonly used housekeeping genes, glyceraldehyde-3-phosphate dehydrogenase, 60S ribosomal protein L27 (RPL27), ornithine decarboxylase antizyme 1, ribosomal protein large P0 (RPLP0) and 40S ribosomal protein S29 (RPS29) were used to normalise the results of target genes. The stability of the expression levels of housekeeping genes was analysed using geNorm( Reference Vandesompele, De Preter and Pattyn 30 ) and NormFinder( Reference Andersen, Jensen and Orntoft 31 ) software packages, as described in their manuals. The RPLP0 and RPL27 genes were selected as the most stable internal pair of controls for normalisation. The information regarding sequence of primers (including annealing temperatures), GenBank accession numbers, PCR efficiency, regression coefficient and span exons for PCR products is provided in Table 2.
Table 2 Characterisation of the select genes used in the real-time quantitative PCR assay

PCR efficiency was calculated for each amplicon using StepOnePlus PCR System software (Applied Biosystems), by amplifying 5-fold serial dilutions of pooled cDNA and run in triplicate. All primer sets exhibited an efficiency ranging from 90 to 110 % and correlation coefficients were higher than 0·99. RT-qPCR reactions were carried out using MicroAmp Optical ninety-six-well plates (Applied Biosystems) in a StepOnePlus thermocycler (Applied Biosystems) in standard cycling conditions. The 12·5-µl PCR reaction mixture contained 6·25 µl of 2×Power SYBR Green PCR Master Mix (Applied Biosystems), 160 nm of forward and reverse primers and 2 µl of diluted cDNA as template. No transcription and no template samples were used as controls. The primer specificity and the formation of primer–dimers were confirmed by melt curve analysis and agarose gel electrophoresis. All the analyses were performed in duplicate, and the relative amounts for each target gene were calculated using the geometric mean of RPLP0/RPL27 as normaliser. The relative expression levels were calculated as a variation of the Livak method( Reference Livak and Schmittgen 32 ), corrected for variation in amplification efficiency, as described by Fleige et al. ( Reference Fleige, Walf and Huch 33 ).
Statistical analysis
All data were checked for normality and variance homogeneity. As variance heterogeneity was detected for most of the variables, data were analysed using the Proc MIXED of SAS software package( 34 ) (version 9.2; SAS Institute). The statistical model included genotype, diet and their respective interaction, as fixed effects, and the repeated statement considering the group option to accommodate the variance heterogeneity. The level of significance was set at P<0·05. The need for covariate adjustment was exploited using hepatic total lipids. Whenever a covariate for each variable was needed, the structure of the covariate model was determined according to the procedures described by Milliken & Johnson( Reference Milliken and Johnson 35 ) and ranged from a simple slope model to individual slopes for each diet×genotype combination. The adjusted variables and their covariance models are identified in the footnotes of tables. As large differences in covariate ranges were intrinsically associated with each genotype, the variable was adjusted and compared with the mean covariate value of each genotype( Reference Milliken and Johnson 35 ). When significant effects were detected, least square means (LSMEANS) were determined using the LSMEANS option and compared using the probability difference procedure adjusted for multiple comparisons, using the Tukey–Kramer method.
Pearson’s correlation coefficients were calculated using the PROC CORR procedure of SAS. A principal component analysis (PCA) was performed with individual fatty acids from the liver. The PRINCOMP procedure was applied to a data set of twenty-four samples and thirty-three variables to reduce the dimensionality of the data set and to describe the variability of data into two dimensions. After data normalisation, the principal components (PC) were considered significant if they contributed to >5 % of the total variance.
Results
Growth performance parameters
Data on growth performance parameters in lean and fatty pigs fed NPD or RPD are shown in Table 3. Fatty pigs had a higher average daily feed intake (ADFI) compared with lean pigs (P<0·001). The inverse was found for gain:feed (G:F) (P<0·001). The RPD, relative to NPD, increased ADFI (P=0·010) but decreased G:F (P<0·001). The average daily gain (ADG) was not affected by genotype or by diet (P>0·05).
Table 3 Effect of the reduced protein diet (RPD) on growth performance variables and plasma biochemical metabolites in lean and fatty pigs (Mean values with their standard errors)

Lean, commercial cross-bred pigs (50 % Large White, 25 % Landrace and 25 % Pietrain); fatty, Alentejana purebred pigs; NPD, normal protein diet; ADFI, average daily feed intake; ADG, average daily gain; G:F, gain:feed; HOMA-IR, homoeostasis model assessment using the insulin resistance index; ALT, alanine aminotransferase (EC 2.6.1.2); AST, aspartate aminotransferase (EC 2.6.1.1); ALP, alkaline phosphatase (EC 3.1.3.1); GGT, γ-glutamyltransferase (EC 2.3.2.13).
a,b,c Mean values within a row with unlike superscript letters were significantly different (P<0·05).
* HOMA-IR, insulin resistance index=(fasting plasma glucose)×(fasting plasma insulin)/22·5.
Plasma biochemical profile
The plasma metabolites are presented in Table 3. The lipid profile was extensively influenced (five out of eight) by genotype. Total lipids (P<0·001), phospholipids (P=0·002), total cholesterol (P=0·002) and LDL-cholesterol (P<0·001) levels were higher in lean than in fatty pigs, in contrast to HDL-cholesterol (P=0·021). Diet only influenced three out of the eight plasma lipids assessed. In fact, the RPD, relative to the NPD, increased TAG (P=0·011), FFA (P=0·026) and VLDL-cholesterol (P=0·009).
Insulin and leptin were affected by genotype, but not by diet, with higher contents in fatty pigs when compared with lean ones (P=0·006 and P<0·001 for insulin and leptin, respectively). Glucose and HOMA-IR were unchanged across experimental groups (P>0·05). Urea levels were lower in both lean pigs (P<0·001) and pigs fed RPD (P=0·001). In addition, a significant interaction between genotype and diet was observed for total protein (P=0·004), with lower values in lean pigs fed RPD.
Regarding the hepatic enzymes, fatty pigs had higher levels of ALT (P=0·040) and GGT (P<0·001) and lower levels of ALP (P<0·001) compared with lean pigs. No influence of diet was found for any of these hepatic markers (P>0·05).
Hepatic total lipids and fatty acid composition
The hepatic lipid content and composition are presented in Table 4. Concerning total lipid content, a genotype effect (P=0·049) was observed, with higher values in the fatty genotype, without influence of dietary treatment.
Table 4 Effect of the reduced protein diet (RPD) on total lipids (g/100 g liver), fatty acid composition (% total fatty acids), partial sums of fatty acids and related ratios in the liver of lean and fatty pigs (Mean values with their standard errors)

Lean, commercial cross-bred pigs (50 % Large White, 25 % Landrace and 25 % Pietrain); fatty, Alentejana purebred pigs; NPD, normal protein diet; G×D, interaction between genotype (G) and diet (D); LC-PUFA, long-chain PUFA.
a,b,c Mean values within a row with unlike superscript letters were significantly different (P<0·05).
* Variable adjusted for total lipids.
† 18 : 1 trans (18 : 1t) represents the sum of 18 : 1 trans6 to trans11. Variable adjusted for total lipids×G×D interaction.
‡ Variable adjusted for total lipids×G interaction.
§ Variable adjusted for total lipids×D interaction.
|| Unidentified minor fatty acids and the dimethylacetals 16 : 0, 18 : 0 and 18 : 1, which are derived from plasmalogens.
¶ SFA=14 : 0+15 : 0+16 : 0+17 : 0+18 : 0+20 : 0+22 : 0+23 : 0.
** MUFA=16 : 1cis-7+16 : 1cis-9+17 : 1cis-9+18 : 1t+18 : 1cis-9+18 : 1cis-11+20 : 1cis-11.
†† PUFA=18 : 2n-6+18 : 3n-3+20 : 2n-6+20 : 3n-3+20 : 3n-6+20 : 3n-9+20 : 4n-6+20 : 5n-3+22 : 4n-6+22 : 5n-3+22 : 6n-3.
‡‡ n-6 PUFA=18 : 2n-6+20 : 2n-6+20 : 3n-6+20 : 4n-6+22 : 4n-6.
§§ n-6 LC-PUFA=20 : 4n-6+22 : 4n-6.
|||| n-3 PUFA=18 : 3n-3+20 : 3n-3+20 : 5n-3+22 : 5n-3+22 : 6n-3.
*** n-3 LC-PUFA=20 : 5n-3+22 : 5n-3+22 : 6n-3.
The predominant fatty acids found across experimental groups were 18 : 0 (28–31 %), 18 : 2n-6 (15–20 %), 16 : 0 (15–16 %), 20 : 4n-6 (11–14 %) and 18 : 1c9 (11–13 % of total FAME). The genotype affected thirteen out of the twenty-seven fatty acids identified. The proportions of 16 : 1c7 (P<0·001), 18 : 1c11 (P<0·001), 14 : 0 (P=0·001), 16 : 1c9 (P=0·001), 20 : 6n-3 (P=0·001), 18 : 3n-6 (P=0·017) and 20 : 5n-3 (P=0·047) were higher in lean pigs, when compared with fatty ones. In contrast, the percentages of 18 : 2n-6 (P<0·001), 20 : 2n-6 (P<0·001), 20 : 3n-6 (P<0·001), 20 : 3n-3 (P=0·001), 20 : 3n-9 (P=0·002) and 23 : 0 (P=0·004) were higher in fatty than in lean pigs. The RPD affected six individual fatty acids. The proportions of 14 : 0 (P=0·009), 18 : 1c11 (P=0·009), 20 : 1c11 (P=0·009), 16 : 1c9 (P=0·030) and 18 : 1c9 (P=0·032) were higher in pigs fed RPD when compared with pigs fed NPD, in contrast with the proportion of 20 : 0 (P=0·014). A significant interaction between genotype and diet was observed for 18 : 1t (P=0·032), with lower values of this minor fatty acid (<0·3 % of total fatty acids) in fatty pigs fed NPD.
Regarding fatty acid sums and ratios (Table 4), the differences observed reflected the variations described earlier for the major individual fatty acids. The genotype influenced both n-6:n-3 ratio (P<0·001) and n-6 PUFA sum (P=0·046), with higher values found in fatty pigs. The proportion of MUFA (P=0·019) was increased by RPD when compared with NPD.
Principal component analysis
A PCA using hepatic fatty acid composition was performed to describe variability of the pooled data into two dimensions (Fig. 1(a)). The score plot of the first two PC explained 51·9 % of the total variability, with 33·7 % for PC1 and 18·2 % for PC2 (Table 5).

Fig. 1 Loading plot of the first and second principal components (PC) of the pooled data (A) and component score vectors (B) for hepatic fatty acid composition (µmol/g liver) from lean (commercial cross-bred pigs (50 % Large White, 25 % Landrace and 25 % Pietrain)) and fatty (Alentejana purebred) pigs fed normal protein diet (NPD) and reduced protein diet (RPD). ![]() , Lean-NPD;
, Lean-NPD; ![]() , lean-RPD;
, lean-RPD; ![]() , fatty-NPD;
, fatty-NPD; ![]() , fatty-RPD.
, fatty-RPD.
Table 5 Loadings for the first two principal components (PC)
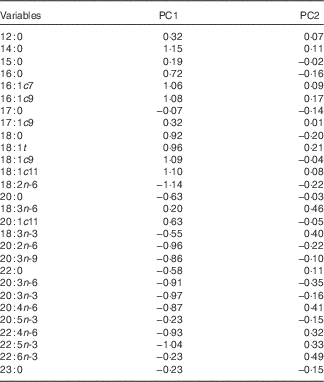
The PC1 was characterised by variables with positive loadings, such as 14 : 0, 18 : 1c11, 18 : 1c9, 16 : 1c9, 16 : 1c7, 18 : 1t, 18 : 0, 16 : 0 and 20 : 1c11, and by variables with negative loadings, such as 18 : 2n-6, 22 : 5n-3, 20 : 3n-3, 20 : 2n-6, 22 : 4n-6, 20 : 3n-6, 20 : 4n-6, 20 : 3n-9, 20 : 0, 22 : 0, 18 : 3n-3, 20 : 5n-3 and 22 : 6n-3 (Table 5). Concerning the PC2, all variables had small contributions with loadings varying between −0·5 and 0·5. The score plot depicted in Fig. 1(b) shows the location of the four experimental groups, lean and fatty pigs fed NPD and RPD, in the multivariate space of the first two PC. These scores were notably arranged in one cluster, corresponding to fatty pigs. The fatty genotype was located mainly in quadrant d, whereas the lean genotype was dispersed across quadrants a and c. The discrimination of dietary treatments was unattainable.
Gene expression levels of key lipogenic enzymes and associated transcription factors in the liver
The expression levels of key genes controlling lipid metabolism in the liver are presented in Fig. 2. Relative ACACA (P<0·001), FASN (P<0·001), SCD (P<0·001), sterol regulatory element-binding factor 1 (SREBF1) (P<0·001) and CRAT (P=0·006) mRNA levels were higher in fatty pigs when compared with lean pigs. In addition, the expression levels of genes were not affected by dietary treatments, except for SREBF1 (P=0·023), with lower values in pigs fed RPD. An interaction between genotype and diet was observed for both FADS1 (P=0·031) and FABP4 (P=0·032) mRNA levels. The RPD increased FADS1 and FABP4 expression levels in lean and fatty pigs, respectively.

Fig. 2 Effect of the reduced protein diet (RPD) on gene expression levels in the liver of lean (commercial cross-bred pigs (50 % Large White, 25 % Landrace and 25 % Pietrain)) and fatty (Alentejana purebred) pigs: (A) acetyl-CoA carboxylase α (genotype, P<0·001), (B) CCAAT/enhancer-binding protein α, (C) carbohydrate response element-binding protein, (D) carnitine O-acetyltransferase (genotype, P=0·006), (E) fatty acid desaturase 1 (FADS1) (genotype×diet, P=0·031), (F) fatty acid desaturase 2 (FADS2), (G) fatty acid-binding protein 4 (FABP4) (genotype×diet, P=0·032), (H) fatty acid synthase (FASN) (genotype, P<0·001), (I) PPARα, (J) stearoyl-CoA desaturase (SCD) (genotype, P<0·001) and (K) sterol regulatory element-binding factor 1 (SREBF1) (genotype, P<0·001; diet, P=0·023). Values are means, with their standard errors represented by vertical bars. a,b,c Mean values within a row with unlike letters were statistically different (P<0·05). ‘Genotype’, ‘diet’ and ‘genotype×diet’ mean significant effect of genotype, diet or interaction between genotype and diet, respectively. For FADS1 and FADS2, the variables were adjusted for total lipids×genotype×diet interaction. NPD, normal protein diet.
Correlation between hepatic fatty acid composition and gene expression levels
The correlation coefficients (r) between fatty acids and gene expression levels in the liver, adjusted for total lipids as covariate, are shown in Table 6. The genes involved in lipogenesis, such as FADS1, FADS2 and FASN, were the most correlated with fatty acid composition, along with SCD, SREBF1 and ACACA.
Table 6 Pearson’s correlation coefficients between fatty acid composition (µmol/g liver) and gene expression levels (relative mRNA level) in the liver of lean and fatty pigs fed normal and reduced protein diets

ACACA, acetyl-CoA carboxylase α; CEBPA, CCAAT/enhancer-binding protein α; ChREBP, carbohydrate response element-binding protein; CRAT, carnitine O-acetyltransferase; FADS1, fatty acid desaturase 1; FADS2, fatty acid desaturase 2; FABP4, fatty acid binding protein 4; FASN, fatty acid synthase; SCD, stearoyl-CoA desaturase; SREBF1, sterol regulatory element-binding factor 1.
Significant correlation: * P<0·05, ** P<0·01, *** P<0·001.
FADS1 mRNA levels were negatively correlated with 18 : 2n-6 (P<0·001), 20 : 2n-6 (P<0·001), 20 : 3n-6 (P<0·001), 20 : 3n-3 (P<0·001), 23 : 0 (P<0·01), 20 : 0 (P<0·05) and 20 : 3n-9 (P<0·05), but positively correlated with 14 : 0 (P<0·001), 16 : 1c9 (P<0·01) and 18 : 3n-6 (P<0·05). FADS2 mRNA levels were correlated with the same fatty acids as FADS1, and also positively correlated with 18 : 1t (P<0·001). The expression levels of FASN gene showed moderate (0·3>r<0·7) negative correlations with 16 : 1c7 (P<0·001), 18 : 1t (P<0·001), 18 : 1c11 (P<0·01), 18 : 3n-6 (P<0·01), 22 : 6n-3 (P<0·01) and 20 : 5n-3 (P<0·05). In addition, FASN mRNA levels were positively correlated with 20 : 3n-6 (P<0·001), 20 : 2n-6 (P<0·05) and 20 : 3n-9 (P<0·05). The SCD relative mRNA levels were negatively correlated with 16 : 1c7 (P<0·001), 18 : 1t (P<0·001), 18 : 1c11 (P<0·01), 18 : 3n-6 (P<0·01), 20 : 5n-3 (P<0·01) and 22 : 6n-3 (P<0·05) and positively correlated with 20 : 3n-6 (P<0·01) and 20 : 3n-3 (P<0·05). SREBF1 mRNA levels showed negative correlations with 16 : 1c7 (P<0·001), 18 : 1c11 (P<0·001), 18 : 1t (P<0·01), 18 : 3n-6 (P<0·05) and 22 : 6n-3 (P<0·05) and a positive correlation with 20 : 3n-6 (P<0·001). ACACA gene expression levels were negatively correlated with 16 : 1c7 (P<0·01), 18 : 1t (P<0·01), 18 : 1c11 (P<0·01) and 22 : 6n-3 (P<0·05) but positively correlated with 20 : 3n-6 (P<0·01). The CRAT gene expression was positively correlated with 20 : 3n-6 (P<0·01), 18 : 0 (P<0·05), 18 : 1c9 (P<0·05), 18 : 2n-6 (P<0·05) and 20 : 3n-3 (P<0·05) fatty acids, and negatively correlated with 18 : 1t (P<0·001).
Discussion
To the best of our knowledge, the present study is the first report on the influence of pig’s genotype (50 % Large White, 25 % Landrace and 25 % Pietrain cross-bred pigs, a lean genotype, v. Alentejana purebred pigs, an autochthonous fatty genotype) and dietary protein levels (normal v. 25 % reduced, not equilibrated for lysine) on hepatic fatty acid metabolism. In order to elucidate the molecular mechanisms of hepatic lipid metabolism in pigs under dietary protein restriction, the expression levels of genes encoding for key lipogenic and lipolytic enzymes and associated transcription factors were also assessed. Moreover, results on the present pig trial regarding the effect of genotype and dietary protein level on fat partitioning between adipose tissue and muscle, as well as the molecular mechanisms regulating the differential fat deposition between those tissues, are published elsewhere( Reference Madeira, Pires and Alfaia 5 ). In brief, dietary protein reduction promoted a lipogenic effect in adipose tissue for both lean and fatty genotypes, but in muscle the lipogenic effect was only observed for lean pigs.
Fatty pigs showed higher ADFI but lower G:F values than lean pigs. This difference is likely due to the higher energy value of body weight gain in fatty pigs. However, ADG was unaffected by both genotype and diet. In contrast, low-protein( Reference Teye, Sheard and Whittington 36 ) and lysine-deficient( Reference O’Connell, Lynch and O’Doherty 37 ) diets decreased ADG in lean pigs during the growing phase. Detailed information about pigs’ performance and feed efficiency is available in the study by Madeira et al. ( Reference Madeira, Costa and Alfaia 38 ).
Regarding genotype, total lipids, phospholipids, total cholesterol and LDL-cholesterol in plasma were higher in lean pigs, whereas HDL-cholesterol was higher in fatty pigs. This finding might be explained by the higher energy needed for the greater protein biosynthesis rate in lean pigs. Delta5 and delta6 desaturases are required for the synthesis of long-chain PUFA that are mainly esterified into phospholipids. These enzymes are widely expressed in mammalian tissues, with the highest levels found in the liver( Reference Nakamura and Nara 12 ). Cholesterol is partially obtained from the diet, through the consumption of animal-derived products, and from de novo biosynthesis in the liver( Reference O’Hea and Leveille 39 ). In the present study, the higher levels of LDL-cholesterol found in lean pigs, contrasting with lower HDL-cholesterol, might result on slightly higher proportions of n-3 PUFA (P=0·052) in the liver.
For hepatic markers, ALT and GGT activities were higher in fatty pigs, whereas ALP activity was inversely higher in lean pigs. Despite the increase in ALT and GGT activities in the fatty genotype, it is worth noticing that the levels found are still within the reference values for pigs, which are 31–58 and 10–52 U/l, respectively( Reference Jackson and Cockcroft 40 ). The restriction of dietary protein did not affect plasma hepatic markers or total lipids, but increased TAG, VLDL-cholesterol and FFA. This finding is consistent with the higher values obtained for total fatty acids in subcutaneous adipose tissue of lean and fatty pigs fed RPD( Reference Madeira, Pires and Alfaia 5 ). In fact, VLDL are produced in the liver and transport endogenous TAG and cholesterol, which are removed from the blood stream for storage in adipose tissue through lipoprotein lipase action.
Insulin levels were higher in fatty pigs, which can be explained by the higher proportion of hepatic n-6 PUFA found in this genotype( Reference Patterson, Wall and Fitzgerald 41 ). In fact, previous studies have shown that young pigs fed low-protein diets in comparison with high-protein diets produced lower glucose and insulin systemic levels( Reference Atinmo, Baldijao and Pond 42 , Reference Caperna, Steele and Komarck 43 ). Regarding the possibility of insulin resistance occurrence, Blat et al. ( Reference Blat, Morise and Sauret 44 ) reported that piglets receiving high-protein diets had higher insulin concentrations, and concomitantly higher HOMA-IR, compared with piglets receiving the NPD. Nevertheless, the values found for insulin resistance index were, once again, within the normal physiological range, that is, <2·4( Reference Blat, Morise and Sauret 44 ). In our study, only fatty pigs fed the NPD had an HOMA-IR above this reference value. Insulin stimulates fatty acid synthesis in the liver with formation and storage of TAG( Reference Wilcox 45 ). This might be the justification for higher total lipids found in the liver of fatty pigs.
Leptin and urea were influenced by genotype, with higher levels of both variables in fatty pigs. Leptin is almost exclusively secreted by adipocytes( Reference Savage and O’Rahilly 46 ), which could explain the higher leptin concentrations found in the fatty genotype. FABP4 plays a key role in fatty acid transport and oxidation, and increases synergistically with leptin during adipose tissue inflammation( Reference Gan, Liu and Cao 47 ). A significant interaction between genotype and diet was observed for FABP4 expression level, with an increase in fatty pigs fed RPD when compared with the lean ones. This finding also agrees with higher leptin levels found in the fatty genotype.
The higher level of urea found in plasma of fatty pigs, relative to lean pigs, may rely on the genetic selection towards higher amino acid deposition, and therefore lower urea excretion. The same effect has been demonstrated in genetically obese rats( Reference Herrero, Remesar and Arola 48 ). In contrast with other reports( Reference Matthews, Southern and Pontif 49 , Reference Gomez, Lewis and Miller 50 ), the restriction of dietary protein decreased plasma urea levels, thus indicating unaffected renal function. The circumstance that restriction of dietary protein only diminished plasma total protein in lean pigs, but not in fatty pigs, might be related to a higher responsiveness of this genotype towards the reduced protein feeding treatment.
An influence of genotype was observed for hepatic total lipids, with fatty pigs showing higher values than lean pigs. This effect might be explained by higher TAG:phospholipid ratios in fatty pigs, knowing that phospholipid content is highly conserved on cellular membranes( Reference Ntawubizi, Raes and Buys 51 ). In addition, this finding concurs with an up-regulation of SCD in fatty pigs relative to lean pigs, and also confirms the major proportion of fatty acids found in the fatty genotype. Similar results were previously reported for both muscle and subcutaneous adipose tissue( Reference Madeira, Pires and Alfaia 5 ). The up-regulated expression of ACACA and SREBF1 genes observed in fatty pigs, when compared with lean pigs, may represent an increased capacity for de novo hepatic lipogenesis. Earlier studies have recognised a regulatory role for transcriptional factors in the liver, fat depots or both tissues in pigs( Reference Kersten, Seydoux and Peters 52 – Reference Cheng and Mukherjee 54 ). The de novo lipogenesis is strongly promoted by SREBF1 activity (most of the genes encoding enzymes involved in fatty acid synthesis are SREBF1c targets), as this transcriptional regulator induces transcription of genes encoding for ACACA, FASN and SCD enzymes, all of them responsible for fatty acid synthesis( Reference Horton, Golstein and Brown 7 ).
Conversely, the restriction of dietary protein had no effect on hepatic total lipids, which concur, following the above described reasoning, with the non-variation of the lipogenic enzyme SCD. It was previously reported that feeding low-protein diets to pigs does not affect backfat thickness, although a small increase in total fatty acid content in both lean (4 %) and fatty (6 %) genotypes was observed in subcutaneous adipose tissue( Reference Madeira, Pires and Alfaia 5 ). However, this increase was not followed by an up-regulation of the expression of the lipogenic enzyme SCD in subcutaneous adipose tissue with RPD( Reference Madeira, Pires and Alfaia 5 ). In contrast, RPD increased IMF content in lean but not in fatty pigs( Reference Madeira, Costa and Alfaia 38 ). The increase in IMF obtained in this pig trial for the lean genotype fed a 25 % RPD (17·5 v. 13·1 % of crude protein) was 40 %. In addition, we have also demonstrated that this lipogenic response in skeletal muscle was due to the limitation of lysine in the diets. The results of mRNA expression in muscle suggest that the genotype-specific effect of RPD on IMF is mediated via up-regulation of the lipogenic enzyme SCD and the adipogenic transcription factor PPARG ( Reference Madeira, Pires and Alfaia 5 ). Therefore, in contrast to muscle (confirmed for lean genotype) and subcutaneous adipose tissue (confirmed for both lean and fatty genotypes), RPD did not promote a lipogenic response in the liver of both genotypes, lean or fatty. This suggests that the restriction of dietary protein does not seem to promote, in the long-term, fatty liver, which is a pathophysiological condition associated with several metabolic disorders, in particular obesity, diabetes and hyperlipidaemia( Reference Lim, Oh and Koh 55 , Reference Kirpich, Marsano and McClain 56 ).
Moreover, the restriction of dietary protein increased MUFA percentages in the liver, as illustrated by 18 : 1c9 fatty acid variations. This result was not corroborated by SCD mRNA expression level, which was unaffected by RPD, as already stated. SCD is the key enzyme required for the biosynthesis of unsaturated fatty acids, which catalyses the 9-cis desaturation of saturated fatty acyl-CoA( Reference Ntambi 57 ). Moreover, in a study using Landrace castrated pigs fed RPD containing plant oils, the SCD-1 protein expression was not affected by these diets in the liver( Reference Dannenberger, Nuernberg and Nuernberg 58 ).
Long-chain PUFA synthesis is dependent on FADS1 and FADS2 desaturases( Reference Nakamura and Nara 12 ). Their coordinated action is supported by our own findings, that is, both enzymes presented significant correlations with the same long-chain PUFA. Moreover, FADS1 mRNA expression was higher in the lean genotype fed the restricted protein diet. In line with this finding, the 20 : 5n-3 fatty acid percentage increased in lean pigs. In addition, the RPD decreased PUFA in muscle and subcutaneous adipose tissue in both genotypes( Reference Madeira, Pires and Alfaia 5 ). In the liver, no significant interactions were observed on fatty acid composition, except for the minor 18 : 1t fatty acids.
Transcriptional regulation is one of the several mechanisms affecting hepatic fatty acid metabolism. Two transcriptional factors, SREBF1 and PPARA, appear to be the key players in the biosynthesis and degradation of fatty acids, respectively( Reference Guillon, Martin and Pineau 59 ). However, this fact was not confirmed in this study for PPARA, whose mRNA levels were similar across experimental groups. Our results showed a clear effect of genotype for SREBF1 with higher mRNA expression levels in fatty pigs rather than in lean pigs. This finding is, once again, probably related to increased plasma insulin, leptin and HDL-cholesterol concentrations in fatty pigs. Insulin is a well-known stimulator of lipogenesis and activates the hepatic expression of SREBF1 ( Reference Osborne 60 ), which in turn is involved in the regulation of genes controlling cholesterol availability( Reference Wang, Sato and Brown 61 ).
The restriction of dietary protein was able to significantly down-regulate SREBF1 mRNA transcriptional levels in both genotypes, suggesting decreased biosynthesis of fat and, most probably, of fatty acid elongation. In line with this, the gene expression levels of ACACA and FASN showed the same trend as that of SREBF1 gene, even if the dietary treatments presented no statistical significance. Previous studies have demonstrated that ACACA and FASN genes are regulated in the liver by other nutritional factors, rather than dietary protein level, such as glucose, fasting/feeding, high-fat and PUFA availability( 18 , Reference Guillon, Martin and Pineau 59 , Reference Flowers and Ntambi 62 ). In a study with Landrace castrate pigs fed RPD supplemented with plant oils, SREBF1c protein expression was not affected by RPD, but ACACA and FASN had decreased protein expressions in the liver( Reference Dannenberger, Nuernberg and Nuernberg 58 ).
As previously reported, fatty acid composition of skeletal muscle and subcutaneous adipose tissue was much more modulated by pig genotype than by dietary protein reduction( Reference Madeira, Pires and Alfaia 5 ). The same finding was observed for hepatic fatty acids. The discriminant analysis presented here, a PCA based on the relationship among hepatic individual fatty acids, shows a clear separation of genotypes. The marked differences between short- and medium-chain MUFA (16 : 1c7, 16 : 1c9 and 18 : 1c11) and long-chain PUFA (20 : 2n-6, 20 : 3n-9, 20 : 3n-6, 20 : 3n-3), as well as 18 : 2n-6 proportions, which characterise the fatty acid profiles of lean and fatty pigs may have contributed for this discrimination. Curiously, for the lean genotype, and in contrast with the fatty genotype, a higher dispersion pattern of cases was observed. One can speculate that higher genetic variability in lean pigs relative to fatty pigs might be responsible for the observed scatter pattern. This remains to be further elucidated.
Conclusions
The present study is the first report on the effects, individual or combined, of restriction of dietary protein level (17·5 v. 13·1 %) and distinct genotypes of pigs (lean v. fatty) on hepatic fatty acid metabolism.
The effect of genotype was the determinant factor for total lipids in plasma and the liver, as well as for hepatic fatty acid composition, because these are two genetic lines of pigs with distinct fat deposition rates. It has been shown that hepatic lipogenesis can be modified in response to genetic selection, as the gene expression levels of key lipogenic enzymes and their associated transcription factors were more affected by genotype than by diet.
The restriction of dietary protein impacted negatively on systemic TAG, FFA and VLDL-cholesterol but promoted no change in total lipids in the liver. Therefore, dietary protein reduction does not seem to enhance fatty acid deposition in the liver, contrary to what has been reported on adipose tissue (both in lean and fatty genotypes) and muscle (only in lean genotype). Thus, these results suggest a tissue-specific lipogenic response of liver to RPD, when compared with adipose tissue and muscle. Ultimately, the restriction of dietary protein does not seem to account for fatty liver development.
However, to a small extent, there is a pig genotype-specific effect of dietary protein restriction on hepatic lipid metabolism, which seems to be mediated by the differential expression levels of FADS1 and FABP4 genes.
Taken together, these data contribute to a better understanding on the molecular mechanisms of dietary protein level on fat partitioning in lean and fatty pigs. The results presented here indicate that the restriction of dietary protein does not promote hepatic lipogenesis in lean or fatty pigs, which is in contrast to the effect described for muscle (only lipogenesis in lean pigs) and adipose tissue (lipogenesis in both lean and fatty pigs). This genotype-specific effect of dietary protein restriction on lipid metabolism in pigs stresses the importance of devising custom-made feeding strategies that take into account the genetic background.
Acknowledgements
The authors are grateful to Eng. J. Santos Silva and Eng. António Sequeira from the Unidade de Investigação em Produção Animal for technical assistance.
The authors acknowledge the financial support from Fundação para a Ciência e a Tecnologia (FCT) grant (PTDC/CVT/99210/2008), CIISA project (UID/CVT/00276/2013) and individual fellowships to M. S. M. (SFRH/BPD/97432/2013) and S. V. M. (SFRH/BPD/63019/2009). V. M. R. P. is an assistant researcher supported through an IF-FCT contract (2013 FCT investigator). P. A. L. is a researcher from the FCT programme ‘Ciência 2008’ and Incentivo 2014 project (AGR/UI0276/2014). This funding contributed to conduct the study and to analyse the samples.
M. S. M., V. M. R. P., C. M. A., P. A. L., S. V. M., R. M. A. P. and J. A. M. P. performed tissue sampling, laboratory work and prepared the manuscript. M. S. M., V. M. R. P., C. M. A., P. A. L. and J. A. M. P. were responsible for the interpretation of the results and preparation of the manuscript. J. A. M. P. was responsible for the study design.
All the authors read and approved the findings of the study. The authors declare that there are no conflicts of interest.











