INTRODUCTION
Lagos bat virus (LBV) constitutes genotype (gt) 2 in the Lyssavirus genus, family Rhabdoviridae [Reference Tordo, Fauquet, Mayo, Maniloff, Desselberger and Ball1]. With the development of molecular biology techniques, the Lyssavirus genus was divided into seven genotypes based on genetic distances. Rabies virus (RABV, gt1) circulates worldwide, whereas LBV, Mokola virus (MOKV, gt3) and Duvenhage virus (DUVV, gt4) have only been isolated from the African continent. European bat lyssaviruses 1 (EBLV-1, gt5) and 2 (EBLV-2, gt6) are present in Europe, and Australian bat lyssavirus (ABLV, gt7) has been identified from the Australian continent. All lyssavirus genotypes have been reported to be pathogenic for animals, and with the exception of gt2, were also reported to cause encephalitis in humans. Four new lyssaviruses, currently listed as tentative species in the Lyssavirus genus, were isolated from bats in Eurasia, i.e. Irkut (IRKV) [Reference Botvinkin2], Aravan (ARAV) [Reference Arai3, Reference Kuzmin4], Khujand (KHUV) [Reference Kuzmin4] and West Caucasian bat virus (WCBV) [Reference Botvinkin2]. Based on phylogeny, serological cross-reactivity and peripheral pathogenicity to mice, lyssaviruses were divided into two phylogroups [Reference Badrane5]. Phylogroup I comprised of genotypes 1, 4, 5, 6, 7 (as well as IRKV, ARAV and KHUV). Phylogroup II includes LBV and MOKV. Members of phylogroup I have been shown to be pathogenic for mice when inoculated via the intracerebral (i.c.) and intramuscular (i.m.) routes. Members of phylogroup II were shown to be pathogenic for mice only when inoculated via the i.c. route but not when inoculated i.m. Importantly, however, this result was based on a study of a single isolate of LBV and a single isolate of MOKV [Reference Badrane5]. Members of phylogroup I cross-neutralize each other. The same is true for phylogroup II, but very limited cross-neutralization was shown between phylogroups I and II [Reference Badrane5]. It has been suggested that WCBV could be considered as a representative of an independent phylogroup III in lieu of genetic distance and absence of serological cross-reactivity with both phylogroups I and II members [Reference Hanlon6, Reference Kuzmin7]. Preliminary pathogenicity studies indicated that WCBV was pathogenic for mice when inoculated i.c. but not i.m., as was observed for phylogroup II, but this virus was pathogenic for hamsters and bats when inoculated i.m. [Reference Kuzmin7]. Commercial rabies vaccine strains all belong to gt1 and there is no evidence of their lack of efficacy against any gt1 viruses although they are much less efficacious against the rabies-related lyssaviruses (gt2–gt7) [Reference Nel8]. For example, various rabies vaccines and anti-rabies immune globulins have been shown to fail to protect animals against MOKV, LBV and WCBV [Reference Hanlon6, Reference Tignor and Shope9, Reference Dietzschold10].
The lyssavirus genome codes for five proteins: Nucleoprotein (N), Phosphoprotein (P), Matrixprotein (M), Glycoprotein (G) and the RNA polymerase (L). G is the most important protein for interaction of virions with host cell receptors and for development of humoral immunity [Reference Dietzschold11–Reference Takayama-Ito16]. In addition, the Arg/Lys333 amino acid (positively charged amino acid) in the G protein ectodomain was identified as being essential for the peripheral virulence of RABV [Reference Tuffereau14, Reference Takayama-Ito16]. Previous genetic analysis indicated that the Arg/Lys333 is replaced by an Asp333 in phylogroup II lyssaviruses, probably resulting in their reduced pathogenicity [Reference Badrane5]. Amino-acid (aa) substitutions in antigenic site II (aa 34–42 and aa 198–200) of G protein in RABV result in a reduction of pathogenicity in adult mice when inoculated via the i.m. route [Reference Prehaud17].
When complete N, P, M and G genes of 13 LBV isolates were analysed phylogenetically, the results identified three different lineages (A–C) of LBV [Reference Markotter18]. One of these lineages (lineage A), demonstrated significant sequence diversity, and was suggested as a new lyssavirus genotype [Reference Markotter18]. The present study was designed to compare the pathogenicity of several isolates of LBV that represent two of the previously three identified phylogenetic lineages of this virus [Reference Markotter18], with representatives of RABV and MOKV. The experiments were performed in a murine model, comparing different doses of the viruses and routes of inoculation. Amino-acid substitutions along G protein, previously suggested to be important for peripheral pathogenicity of lyssaviruses, were also compared.
METHODS
Eleven lyssavirus representatives were included in the present study (Table 1). These isolates were amplified in suckling mouse brain using i.c. inoculation. Selection of the LBV representatives was based on their phylogenetic positions in lineages A and C (Fig. 1) and their ability to be amplified to significant titres by mouse inoculation. The single available isolate of lineage B (LBVNIG1956) was not included because titres of this virus were low even after extensive passaging in mouse brain and cell cultures. The presence of lyssavirus antigen in the mouse brains was confirmed by direct fluorescent antibody test (FAT) using FITC-labelled monoclonal anti-rabies globulin (Jujirebio Diagnostics, USA) [Reference Dean, Abelseth, Atanasiu, Meslin, Kaplan and Koprowski19]. Ten percent mouse brain suspensions were prepared in Minimum Essential Medium (MEM-10, Gibco, USA) supplemented with 10% fetal calf serum. The suspensions were centrifuged at 3200 g for 15 min and stored at −80°C. The titre of the inoculum was determined by i.c. inoculation of virus dilutions into 4-week-old outbred ICR mice, and the 50% mouse i.c. lethal dose (MICLD50) was calculated using the Spearman–Karber method [Reference Aubert, Meslin, Kaplan and Koprowski20].
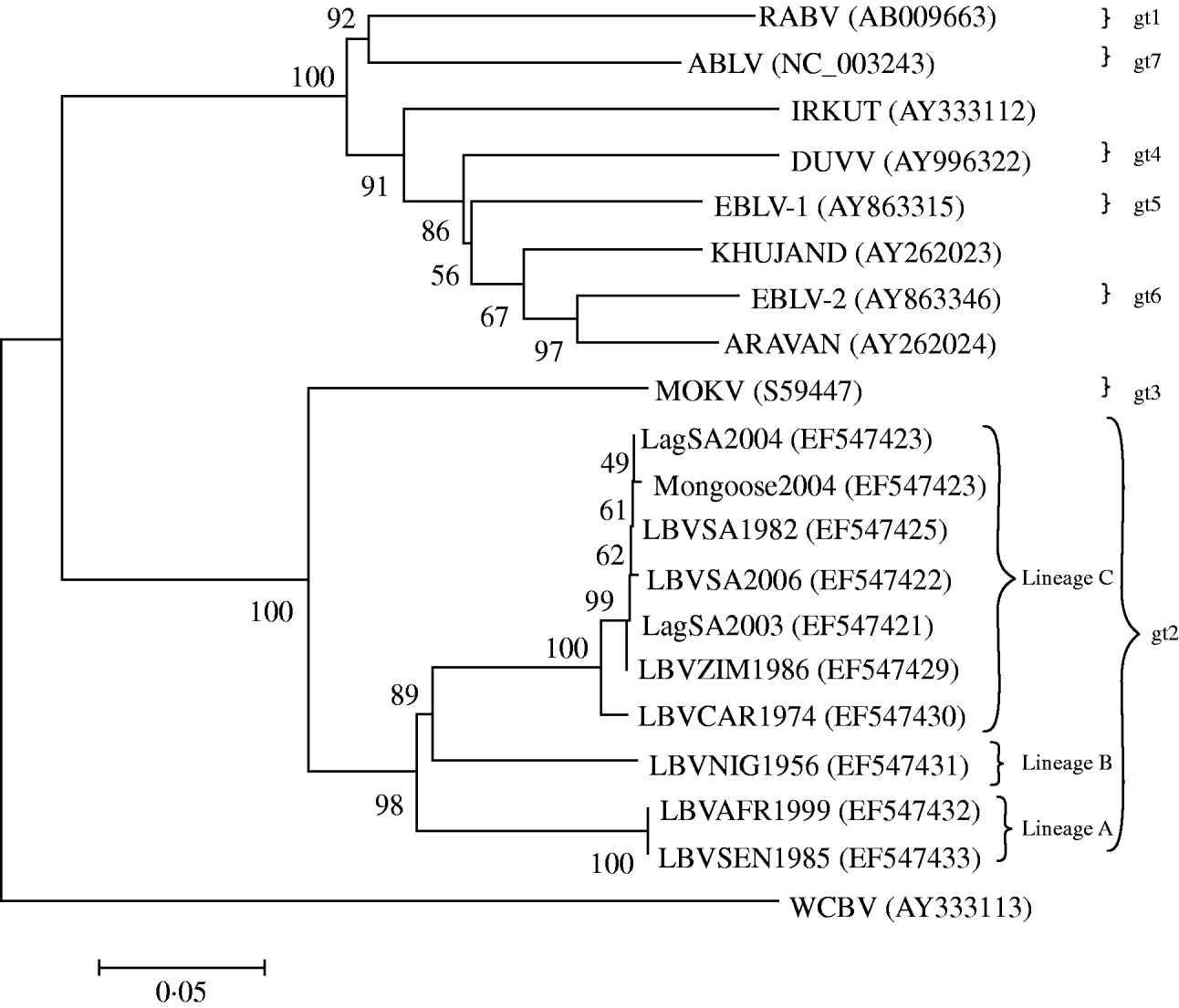
Fig. 1. Neighbour-joining (NJ) phylogenetic tree based upon the 439 amino acids of the ectodomain of the G protein of representatives of the lyssavirus genus, obtained by the NJ method. GenBank accession numbers are indicated for each isolate. Bootstrap values are indicated at the nodes and branch lengths are drawn to scale.
Table 1. Information about lyssavirus isolates used in experimental infections of mice

Four-week-old ICR mice (obtained from Harlan Sprague–Dawley, USA) were used for experimental infections. Each mouse was identified with an ear tag providing a unique number (National Band and Tag Co., USA). All animal care and experimental procedures were performed in compliance with the Centers for Disease Control and Prevention Institutional Animal Care and Use Guidelines (USA). The mice were inoculated with lyssavirus isolates using different routes of inoculation and different doses of inoculum: group A (102 MICLD50 i.c.), group B (103 MICLD50 i.m.), group C (106 MICLD50 i.m.). Each group consisted of five mice. Mice were observed for 56 days and clinical signs and mortality were recorded daily. The i.m. inoculation was performed into the gastrocnemius muscle, in a total volume of 50 μl. The i.c. inoculation was performed in a total volume of 30 μl as described previously [Reference Koprowski, Meslin, Kaplan and Koprowski21]. The FAT was performed on mouse brain collected from succumbed or euthanized mice at the end of the experiment on day 56. The nucleotide sequences of the complete G genes of LBV isolates were generated as described previously [Reference Markotter18]. Nucleotide sequences obtained were assembled and edited using Vector NTI 9.1.0 (Invitrogen, USA) and amino-acid sequences were deduced using the translate function of this program. Multiple sequence alignments were generated using the Clustal X program [Reference Jeanmougin22].
RESULTS
Susceptibility
The i.c. inoculation of mice with gt1–gt3 lyssavirus isolates produced similar pathogenicity profiles, all leading to 100% mortality (Fig. 2). Intramuscular inoculation with gt1–gt3 lyssaviruses produced more variable results. When a high dose (106 MICLD50) was used, all viruses were able to induce disease and subsequent death, but they were not equally virulent. At this dose, the RABV (isolate WAmyotis) and two of the LBV isolates, both from lineage A (LBVSEN1985 and LBVAFR1999), caused 100% fatality rates after i.m. administration of the inoculum. Other LBV isolates injected i.m. at this dose caused rabies in 20–60% of mice. When a representative of gt3 [isolate MOKVSA(252/97)] was inoculated i.m. at this dose, only 20% of the mice succumbed. When viruses were introduced i.m. at a lower dose (103 MICLD50) the virulence decreased, and no mice inoculated with isolates MOKVSA(252/97), LBVCAR1974, LBVSA1982 and LBVSA2006 developed rabies. In contrast, the most significant virulence was observed for isolates LBVSEN1985 and LBVSA2004 (60% and 40%, respectively).
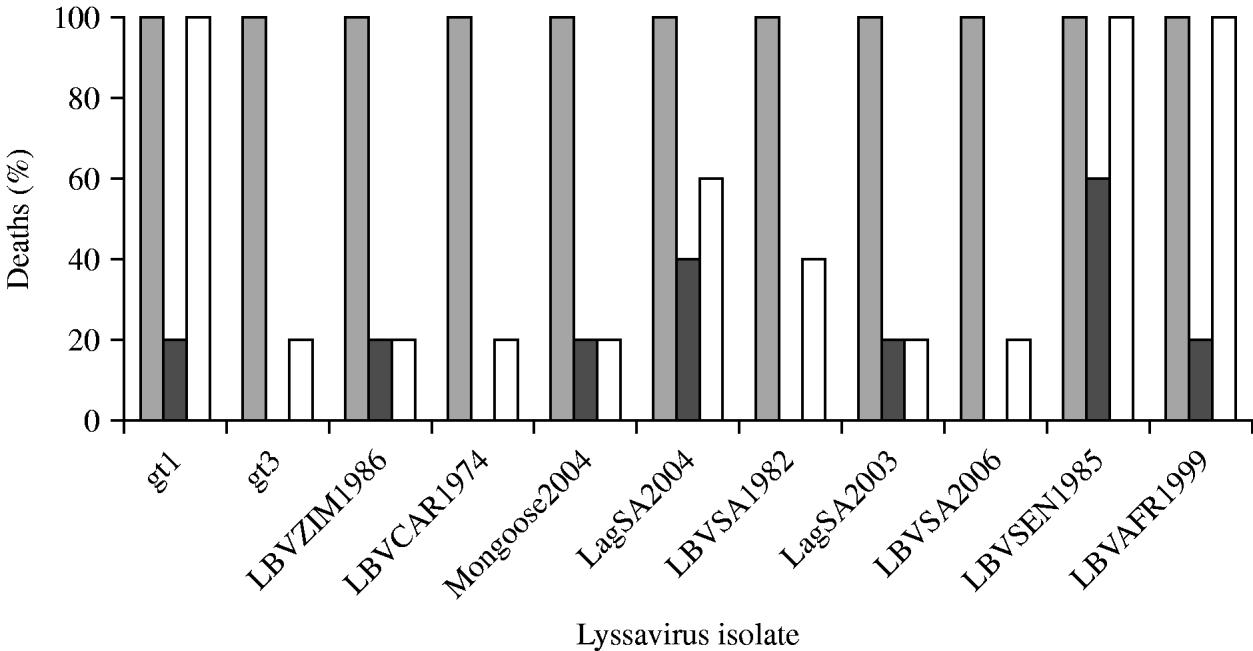
Fig. 2. Virulence of genotype (gt) 1 (WAmyotis), gt2 and gt3 [MOKVSA(252/97)] lyssaviruses in 4-week-old mice after intracerebral (i.c.) and intramuscular (i.m.) inoculation. Results are expressed as a percentage of rabid animals after observation for 56 days. Different viral doses were introduced: (a) ![]() , 102 MICLD50 i.c.; (b)
, 102 MICLD50 i.c.; (b) ![]() , 103 MICLD50 i.m.; (c) □, 106 MICLD50 i.m.
, 103 MICLD50 i.m.; (c) □, 106 MICLD50 i.m.
The duration of incubation periods was also found to be dependent on the specific virus isolate and the route of inoculation, and was proportional to the inoculation dose (Fig. 3). The i.c. inoculation produced the shortest incubation period for all virus isolates tested. RABV (isolate WAmyotis) did not demonstrate a significant difference in the mean incubation periods between 102 MICLD50 i.c. and 106 MICLD50 i.m., but when introduced i.m. at dose of 103 MICLD50, the duration of the incubation period increased to 17 days. For a representative of gt3 [MOKVSA(252/97)], the mean incubation period increased between the i.c. and i.m. inoculations, and only the 106 MICLD50 dose caused the disease by the i.m. route, and in one mouse only. The shortest mean incubation periods were observed for LBVSEN1985 and LBVAFR1999 for all the routes of inoculation and over the entire dosage spectrum.
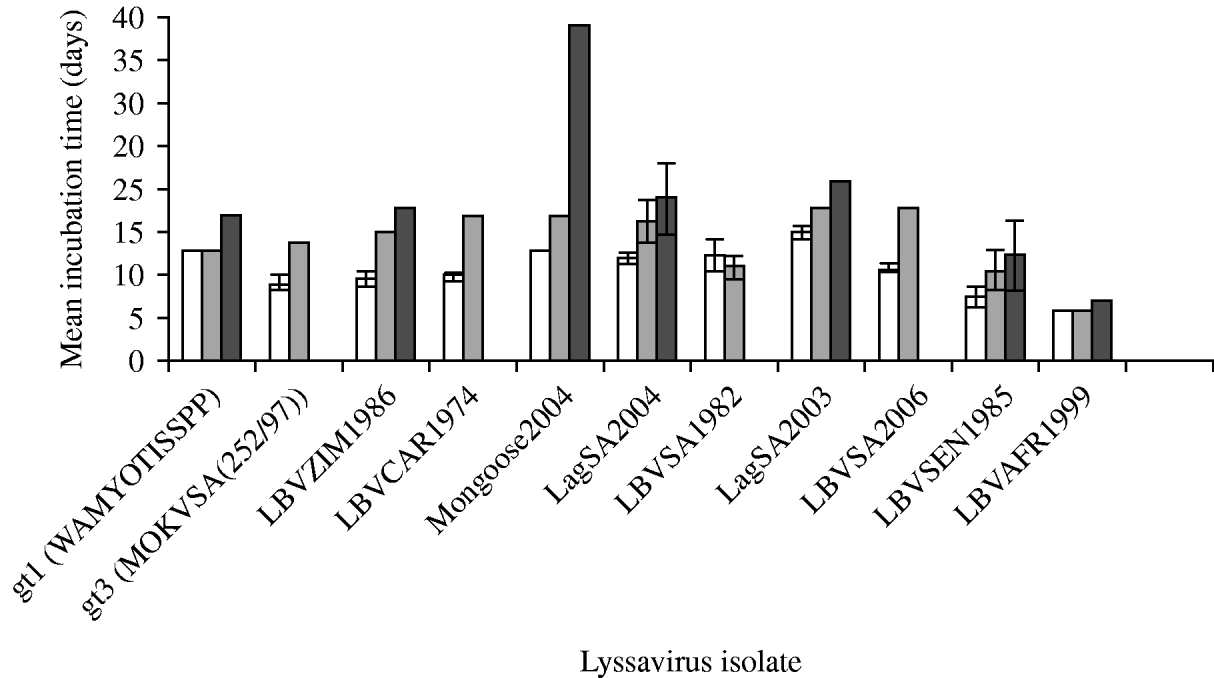
Fig. 3. Mean incubation time (days) of lyssavirus isolates after different routes of inoculation [intracerebral (i.c.) and intramuscular (i.m.)] and different viral doses were introduced into 4-week-old mice. The standard deviation (s.d.) is indicated. Some s.d. values were 0. (a) □, 102 MICLD50 i.c.; (b) ![]() , 106 MICLD50 i.m.; (c)
, 106 MICLD50 i.m.; (c) ![]() , 103 MICLD50 i.m.
, 103 MICLD50 i.m.
Domains of the lyssavirus G that were previously implicated in pathogenesis were compared between viruses used in our study and other representatives (Figs 4 and 5). Amino-acid positions 330 and 333 on the ectodomain were found to be conserved in genotypes 1, 4, 5, 6, 7, ARAV, IRKV and KHUV but not in gt2, gt3 and WCBV. In gt2 and gt3 isolates, Arg/Lys333 was replaced by an aspartic acid (Asp) (Fig. 4). With the exception of two lineages (B and C) of gt2 and WCBV, Lys330 was found to be conserved within the Lyssavirus genus. The isolates belonging to lineages B and C of gt2, were found to possess Leu330. In contrast, isolates belonging to lineage A of gt2 contain Lys330, similar to all other lyssavirus genotypes. A multiple alignment of G sequences of gt2 representatives used in this study is shown in Figure 5. The lineage A isolates (LBVSEN1985 and LBVAFR1999) demonstrated significant sequence diversity in comparison to the other gt2 isolates. Within antigenic site II (positions 34–42 and 198–200), sequence diversity was found at positions 37 and 42 between lineage C isolates and at positions 37, 39, 40, 42, 198 and 200 between the more pathogenic LBV lineage A and the less pathogenic LBV lineage C. Substitutions were also observed in aa 330, 334 and 336 of antigenic site III between lineage A and lineage C isolates. No differences were observed between LBV isolates for other known antigenic sites within G. When anlaysing other sites on the G previously implicated as being significant for pathogenicity (positions 164, 182, 205, 210, 242, 255, 268, 303) only position 205 indicated an amino-acid difference between lineage A and lineage C isolates (Fig. 5).
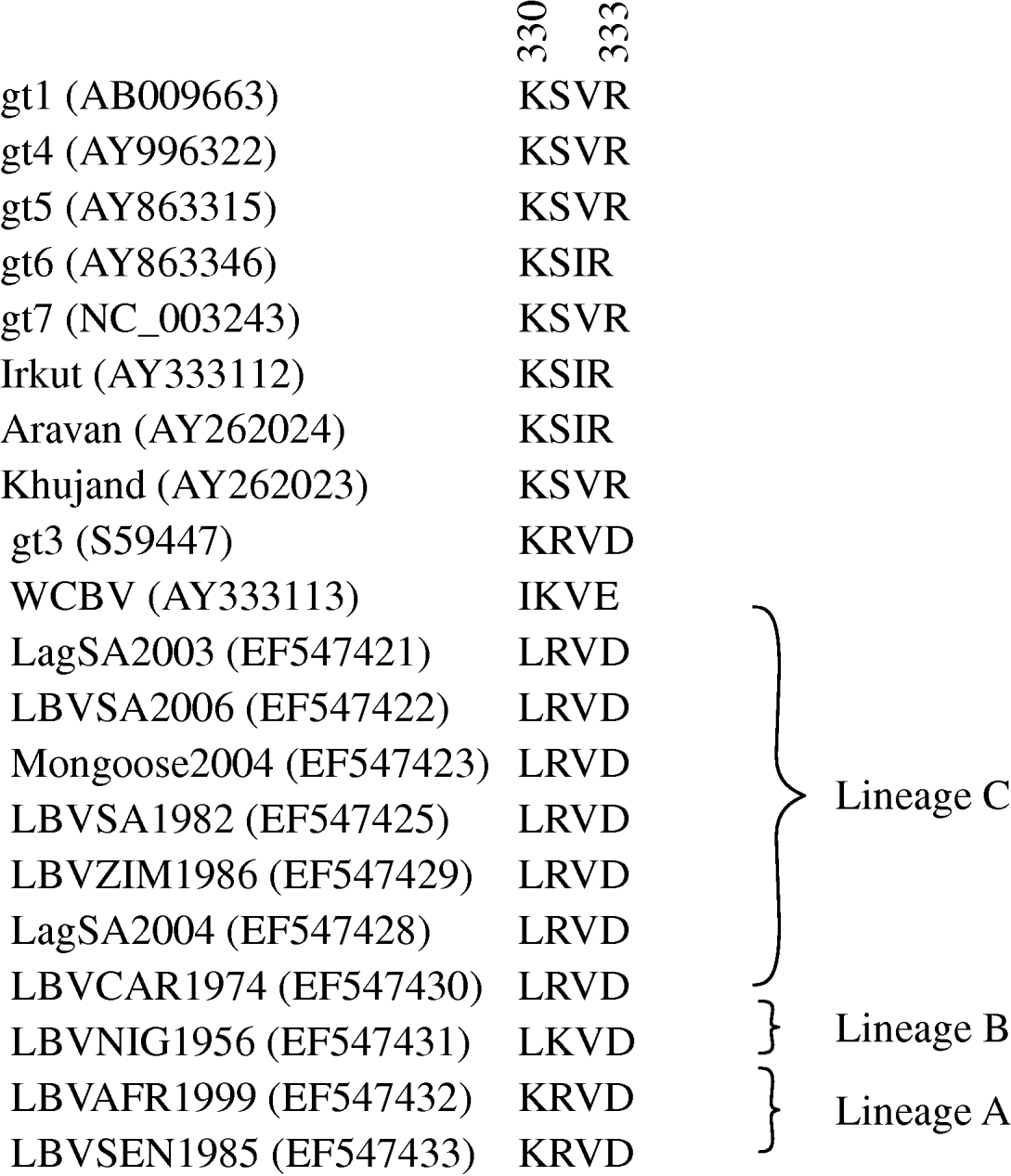
Fig. 4. Multiple alignment indicating amino acids 330–333 of the ectodomain of the G protein of representatives of the lyssavirus genus. GenBank accession numbers of the sequences are indicated.
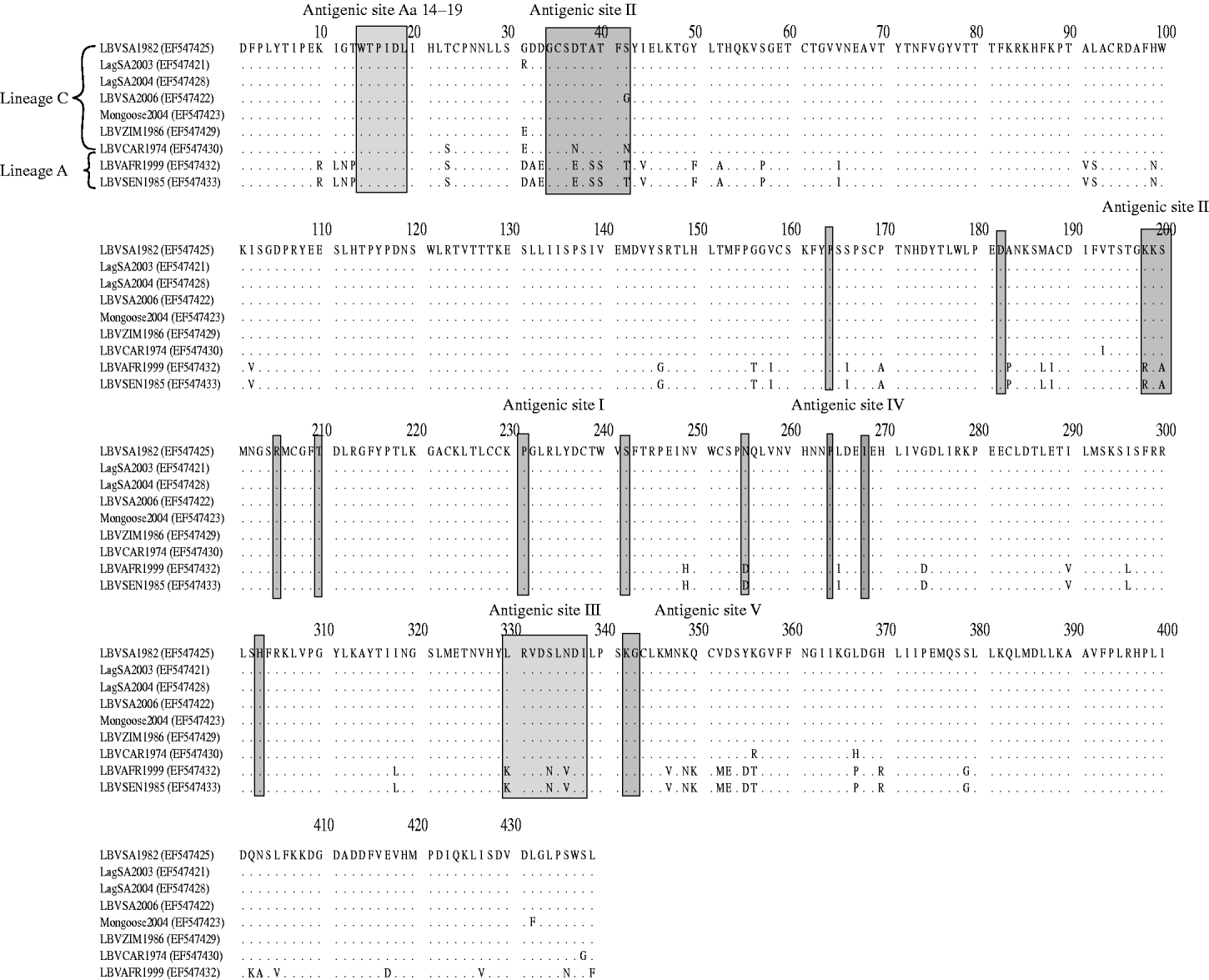
Fig. 5. Multiple alignment indicating differences in the G protein of genotype 2 representatives analysed in pathogenicity studies. Antigenic sites, as well as domains previously indicated as playing a role in pathogenesis are indicated.
DISCUSSION
The aspect of relative pathogenicity, with reference to inoculum dose and route of inoculation, is an important criterion in considerations of lyssavirus ecology. Previous studies suggested that phylogroup II lyssaviruses were not pathogenic when introduced peripherally. These assertions contributed directly to the suggestion that such viruses are generally less pathogenic, and imply that they have limited public health and veterinary significance [Reference Badrane5]. In the present study we have assessed the susceptibility of mice to various isolates that are classified within the lyssavirus gt2 (LBV) in comparison to one isolate each from gt1 (RABV) and gt3 (MOKV). A single isolate of MOKV and RABV (known to be virulent) were included for the purpose of comparison. When inoculated i.c., all lyssaviruses in our panel caused acute progressive encephalitis (rabies). However, differences were observed when these viruses were inoculated i.m. at peripheral sites distant to the central nervous system (CNS).
We have shown that several representatives of LBV caused rabies in mice when introduced i.m. In the case of MOKV that was inoculated i.m. at a dose of 106 MICLD50, only 20% of mice succumbed, whereas the RABV isolate and two LBV isolates were fatal to the entire respective groups of mice, following the same dose and route of infection. Of even more significance was the finding that even at a reduced viral dose (103 MICLD50) inoculated via the i.m. route, six LBV isolates from lineages A and C demonstrated equal or greater pathogenicity to mice than did RABV.
Several studies of the lyssavirus G protein suggested specific epitopes that may be involved in pathogenicity. For example, mutations in the G protein can affect pathogenesis as indicated by the amino-acid replacement at position 333 of the ectodomain. The presence of a positively charged amino acid in this position, Arg or Lys, led to a virulent phenotype of RABV while mutations to a Gln, Ile, Gly, Met or Ser led to a less pathogenic or avirulent virus [Reference Dietzschold11, Reference Seif12, Reference Tuffereau14, Reference Takayama-Ito16]. The aa 333 mutation could also affect the rate of viral spread from cell to cell [Reference Dietzschold23]. A double mutation of aa 330 and aa 333 led to a further reduction of pathogenicity of RABV compared to a single aa 333 substitution [Reference Seif12, Reference Coulon15]. A recombinant virus with a Glu333 reverted back to a more pathogenic phenotype when Asn194 mutated to Lys194 during suckling mouse brain passage [Reference Faber24]. Badrane et al. [Reference Badrane5] reported limited peripheral pathogenicity of phylogroup II lyssaviruses, presumably related to the Asp333 in their glycoprotein ectodomain. From the results of our study it is evident that Asp333 is not the sole determinant of reduced pathogenicity since LBV isolates from lineage A (Asp333) demonstrated the same peripheral pathogenicity to mice as a RABV (Arg333) isolate. The significance of Lys330 within lineage A representatives (similar to the phylogroup I lyssaviruses) distinguishes them from the other LBV lineages, and is difficult to assess. For example, Lys330 is also present in the MOKV isolate that demonstrated reduced peripheral pathogenicity in our study. However, we only analysed one isolate of MOKV, and future studies including different MOKV representatives may provide a better resolution. Mutations in antigenic site II of the G has previously been shown to render a less pathogenic laboratory RABV strain CVS when introduced i.m. into adult mice [Reference Prehaud17]. When comparing the amino-acid sequence of antigenic site II of LBV isolates analysed in this study, differences were observed between isolates of lineages A and C. A difference between isolates of lineages A and C was also observed in aa 255, which has previously been implicated as being essential in the pathogenicity of the virulent Nishigahara strain (gt1) [Reference Takayama-Ito16]. Our results suggest that certain LBV representatives, particularly from phylogenetic lineage A (which was recently suggested for consideration as an independent genotype [Reference Markotter18]), demonstrate the same or even greater peripheral pathogenicity to mice, as a RABV representative and suggest that the pathogenicity of phylogroup II lyssaviruses has been underestimated. Variation in pathogenicity can occur within a genotype, depending on the strain origin and animal model [Reference Constantine25–Reference Baer and Bales29]. Even if a mouse is a standard laboratory species for lyssavirus infection, this is not the natural lyssavirus host, and therefore probably not the best model for detailed pathogenesis studies. Indeed, all available phylogroup II isolates were obtained from naturally infected wild and domestic mammals, indicating that these viruses have well established pathways for natural circulation. Intradermal exposures could in some cases be worthy of consideration since LBV is associated with large fruit bats, e.g. Rousettus, Epomophorus and Eidolon [Reference Markotter18] from which a bite can easily access mammalian muscles. Our findings clearly indicate the need for improved surveillance and public health precautions for phylogroup II lyssaviruses. Considering that commercially available rabies vaccines do not protect against phylogroup II lyssaviruses, new biologicals which would be capable of protecting against such viruses [Reference Hanlon6, Reference Nel8], are needed and have been shown to be feasible, at least experimentally [Reference Bahloul30].
ACKNOWLEDGEMENTS
The authors thank Dr C. T. Sabeta (Agricultural Research Council, Onderstepoort Veterinary Institute, Rabies Unit, South Africa) for providing the MOKVSA(252/97) and LBVSA1982 isolate and Dr F. Cliquet [Agence Française de Sécurité Sanitaire des Aliments (AFSSA), France] for providing the LBV1999AFR isolate. This study was supported in part by the National Research Foundation of South Africa, the University of Pretoria International Affairs Officee's Postgraduate Study Abroad Bursary Programme, and the US National Vaccine Program Office. Use of trade names and commercial sources are for identification only and do not imply endorsement by the U.S. Department of Health and Human Services. The findings and conclusions in this report are those of the authors and do not necessarily represent the views of the funding agencies.
DECLARATION OF INTEREST
None.








