INTRODUCTION
Brachyspira hyodysenteriae causes swine dysentery (SD), a severe mucohaemorrhagic diarrhoeal disease that primarily affects pigs during the growing-finishing period [Reference Hampson, Fellström, Thomson, Straw, Zimmerman, D'Allaire and Taylor1].
With 15% of the total European Union (EU) output, Spain ranked second in terms of EU pork production in 2007 (source: Eurostat). Spanish swine production has grown significantly in recent years, increasing the number of large swine production units raising white commercial breeds under intensive conditions. Moreover, 10% of the sows in Spain belong to an autochthonous breed designated as Iberian pig (source: Spanish Ministry of Environment and Rural and Marine Affairs). This local breed is characterized by its rusticity and has been traditionally reared in extensive units. In recent years, Iberian pigs have also been reared in semi-intensive units in order to make their production more profitable.
SD has been described in all countries with a swine industry and is considered one of the most significant production-limiting porcine infections [Reference Hampson, Atyeo, Combs, Hampson and Stanton2]. In Spain, the importance of SD as a cause of diarrhoea among growers, finishers and sows has been investigated [Reference Carvajal3], with more than 30% of Spanish farms and 12% of faecal specimens testing positive for B. hyodysenteriae. Moreover, decreased susceptibility to the main antimicrobials used in the treatment of SD has been detected in Spanish B. hyodysenteriae isolates [Reference Hidalgo4].
Diverse methodologies, such as serotyping [Reference Hampson5], restriction endonuclease analysis (REA) [Reference Combs, Hampson and Harders6], multilocus enzyme electrophoresis (MLEE) [Reference Trott, Oxberry and Hampson7], pulsed-field gel electrophoresis (PFGE) [Reference Atyeo, Oxberry and Hampson8], random amplified polymorphic DNA (RAPD) [Reference Dugourd9], biochemical characterization [Reference Fellström10], DNA restriction fragment polymorphism analysis [Reference Jensen, Casey and Stanton11] and multilocus sequence typing (MLST) [Reference Råsbäck12], have been used to characterize and analyse the diversity of Brachyspira spp. isolates.
The research reported herein was performed to describe the genetic and phenotypic diversity of Spanish B. hyodysenteriae field isolates and to investigate epidemiological relationships between them. Moreover, we attempted to confirm the presence of tiamulin-resistant isolates and to investigate their common or independent origin.
METHODS
Bacterial strains and growth conditions
A set of 74 Spanish isolates of strongly β-haemolytic intestinal spirochaetes recovered from pigs and classified as B. hyodysenteriae according to species-specific PCR [Reference Leser13] was used in the current study. Isolates were selected in order to include samples representing the most important pig production regions of the country. All isolates were obtained from faecal samples from growers, finishers or sows submitted for routine diagnostics to the Laboratory of Infectious Diseases in the Veterinary Faculty at the University of León, Spain, and stored in liquid nitrogen. A list depicting isolate designation, herd, date of isolation, geographical origin and other relevant information, if available, is presented in Table 1. The isolates were sent in Amies medium to the National Veterinary Institute (SVA), Uppsala, Sweden, where they were tested using duplex PCR [Reference Råsbäck14], based on the tlyA and the 16S rRNA genes, for detection of B. hyodysenteriae and B. pilosicoli, respectively.
Table 1. Isolate designation, herd, date of isolation, geographical origin, RAPD and PFGE patterns and other relevant information, when available, for 74 Spanish B. hyodysenteriae field isolates included in the current study
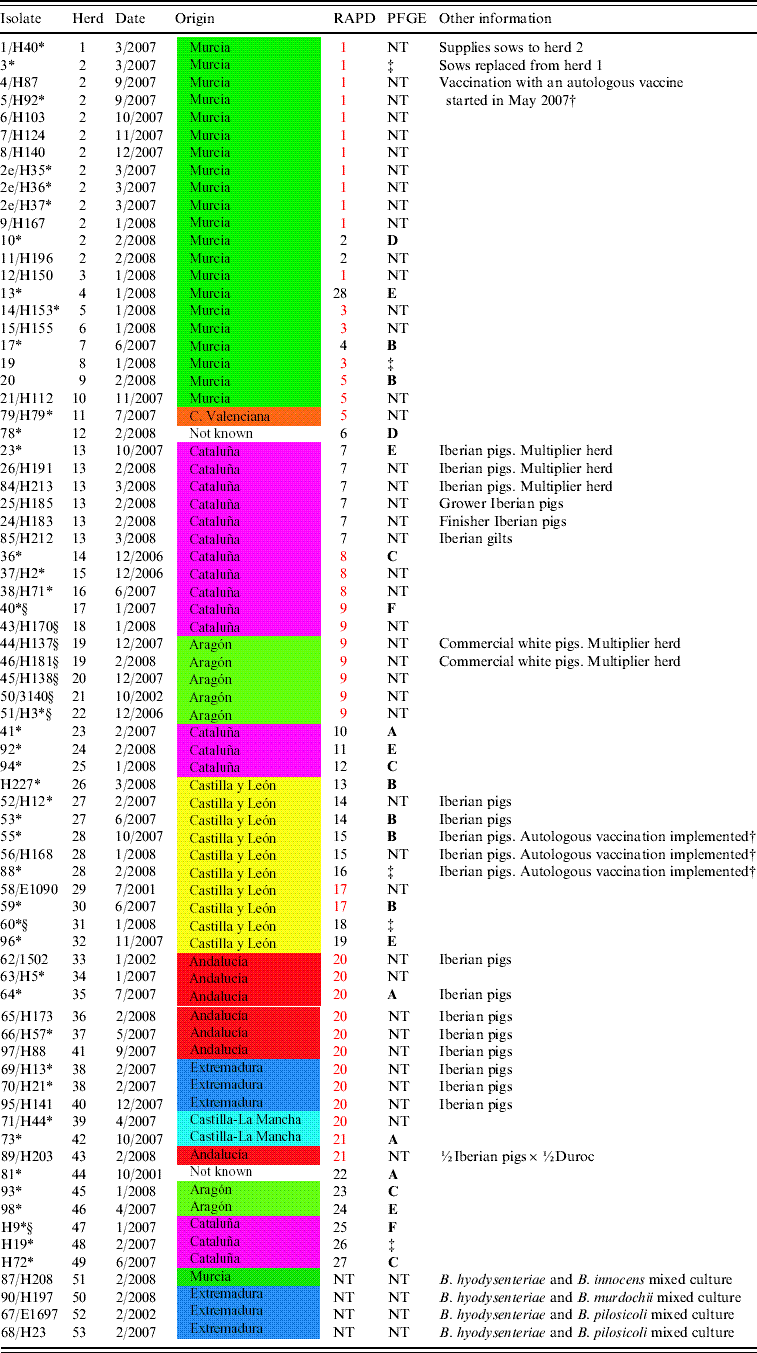
Date: Month and year of isolation; Origin: administrative region, coloured according to the map (right), where the farm was located; RAPD: pattern assigned in the RAPD study (RAPD patterns in red type are shared by isolates from different herds); PFGE: pulsed-field gel electrophoresis cluster for MluI, according to groups (A–F) established in Figure 2.

NT, Not tested.
* Isolates tested with smpA/smpB PCR.
† Autologous B. hyodysenteriae vaccination programme consisting of a whole-herd vaccination repeated each 4 months.
‡ PFGE tested and not clustered with an 80% cut-off value.
§ Indol-negative Spanish B. hyodysenteriae field isolates.
We also investigated two German indole-negative B. hyodysenteriae isolates, designated 5677/96 and T4 [Reference Råsbäck12] from the Swedish collection at SVA and the B. hyodysenteriae reference strain B204 (ATCC 31212), B. hyodysenteriae type strain B78T (ATCC 27164T), and B. pilosicoli type strain P43/6/78T (ATCC 51139) were used as controls for PCR and biochemical characterization.
Bacteria were grown on fastidious anaerobe agar (FAA, SVA, Sweden) at 42°C in anaerobic jars [GENbox (bioMérieux, France) with AnaeroGen sachets (Oxoid, UK)].
Biochemical tests and β-haemolysis
Biochemical characterization was performed as previously described by Fellström & Gunnarsson [Reference Fellström and Gunnarsson15]. In brief, 3-day-old cultures were tested for weak or strong β-haemolysis on trypticase soy agar supplemented with 5% ovine blood. Indole production was investigated using the spot indole test; α-galactosidase activity was determined using diagnostic tablets (Rosco Diagnostica, Denmark) and hippurate hydrolysis as described by Rübsamen & Rübsamen [Reference Rübsamen and Rübsamen16].
Testing antimicrobial susceptibility
Eleven Spanish B. hyodysenteriae isolates (for reference see Table 2), selected on the basis of their reduced susceptibility to tiamulin (⩾2 μg/ml) determined in a previous investigation [Reference Hidalgo4], were tested for antimicrobial susceptibility using VetMIC™ Brachy QCR high panels (SVA, Sweden) according to the manufacturer's protocol. The antimicrobial agents tested were tiamulin, valnemulin, doxycycline, lincomycin, tylosin, and tylvalosin. The minimum inhibitory concentration (MIC) was determined as the lowest concentration of antimicrobial agent that prevented visible growth. Absence of contamination was checked by phase contrast microscopy.
Table 2. Minimum inhibitory concentrations (μg/ml) of six antimicrobial agents for 11 Spanish B. hyodysenteriae field isolates selected on the basis of their reduced susceptibility to tiamulin (⩾2 μg/ml) determined in a previous investigation [Reference Hidalgo4]

RAPD
Seventy B. hyodysenteriae isolates confirmed by duplex PCR [Reference Råsbäck14] and biochemical tests [Reference Fellström and Gunnarsson15] as well as the reference and type strains of B. hyodysenteriae (B204 and B78T) were typed by RAPD following the technique described by Quednau et al. [Reference Quednau17], slightly modified. DNA samples were prepared from 3-day-old pure cultures grown on FAA. Two filled 1-μl loops of the bacteria were washed twice in phosphate buffered saline (pH 7·3), boiled in nuclease-free water (Sigma-Aldrich, USA) and centrifuged. The supernatant was transferred to a sterile microtube. Extracted DNA samples were adjusted to a concentration of 20 ng/μl. RAPD fingerprints were generated with primer P73 (5′-ACGCGCCCT-3′) and primer P1254 (5′-CCGCAGCCAA-3′), resulting in two different pattern sets that were visually analysed. Results were interpreted with strict criteria and isolates which differed in at least one fragment (including weak, barely visible and broad bands) were assigned to different RAPD types. In order to ensure reproducibility, this technique was repeated at least three times for each isolate.
PFGE
Thirty-one B. hyodysenteriae isolates were typed by PFGE, including 28 Spanish field isolates representing the different RAPD patterns (see Table 1), the reference and type strains of B. hyodysenteriae (B204 and B78T), and one German indole-negative isolate (5677/96).
The DNA preparation procedure for PFGE was adapted from a previous protocol described for Treponema spp. [Reference Pringle18]. For each isolate, bacterial cells from two FAA plates were harvested, suspended in 10 ml TE buffer (10 mm Tris, 1 mm EDTA) and washed three times in 5 ml TE buffer. The cells were then suspended in 1·5 ml Pett IV buffer (10 mm Tris–HCl, 1 m NaCl), adjusted to an optical density of 0·800 at 405 nm and mixed 1:1 (v:v) with 1·5% low melting temperature agarose (NA agarose, GE Healthcare, UK). The agarose plugs were incubated in ESP (0·5 m EDTA, 1% N-lauroyl sarcosine, 0·2% pronase E) at 50°C for 24 h, restoring the liquid after 1·5 h. Gel plugs were then washed six times in TE buffer. Digestion with restriction enzymes MluI (5′-A↓CGCGT) and SalI (5′-G↓TCGAC) and pulsed-field electrophoresis were performed as described by Fellström et al. [Reference Fellström10], using a CHEF-DR® III pulsed field electrophoresis system (Bio-Rad Laboratories AB, Sweden) at 6 V/cm2 with an included angle of 120°. Initial and final switch times were 5 s and 70 s, respectively. The gels were run for 24 h in 0·5×TBE buffer (44·5 mm Tris, 44·5 mm boric acid, 1 mm EDTA) at 14°C and subsequently stained with ethidium bromide. A lambda marker (New England, Biolabs, USA) was included to normalize the PFGE banding patterns that were used for producing dendrograms, following calculation of the Dice coefficient and analysis with the unweighted pair-group method by arithmetic averages (UPGMA) clustering fusion strategy, performed with the GelCompar program (Applied Maths, Belgium).
SmpA/smpB-specific PCR
A PCR assay for specific detection of smpA or smpB genes was performed on 42 Spanish B. hyodysenteriae field isolates (listed in Table 1) as described by Holden et al. [Reference Holden19]. Genomic DNA was prepared by the CTAB extraction method and at least one isolate per RAPD pattern was included. Reference strain B204 was included as smpA-positive control.
RESULTS
PCR identification and biochemical characterization
Duplex PCR analysis for the detection of B. hyodysenteriae and B. pilosicoli, resulted in the tlyA gene fragment being amplified for all 74 isolates; the 16S rRNA gene fragment specific for B. pilosicoli was amplified for two isolates (67/E1697 and 68/H28). These latter two isolates were considered to be B. hyodysenteriae and B. pilosicoli mixed cultures. In addition, the biochemical tests placed 70 of the 72 presumptive B. hyodysenteriae isolates (according to the duplex PCR) in group I (B. hyodysenteriae) [Reference Fellström20]. However, isolates 90/H197 and 87/H208 were classified as group III, B. innocens and B. murdochii, respectively, and considered as mixed cultures. Sixty-one group I isolates (87·1%) were recorded as indole positive in the spot indole test, while nine group I isolates (12·9%; 40, 43/H170, 44/H137, 46/H181, 45/H138, 50/3140, 51/H3, 60 and H9), were indole negative.
MIC determinations
The MICs of the six antimicrobial agents studied for the 11 selected Spanish B. hyodysenteriae isolates are shown in Table 2.
RAPD analysis
Twenty-eight dissimilar RAPD patterns were obtained for the 70 Spanish B. hyodysenteriae field isolates. A different figure was given for each RAPD pattern (Table 1). German indole-negative isolates, 5677/96 and T4, were assigned to RAPD pattern number 9. Reference and type strains B204 and B78T did not share any RAPD pattern with the studied field isolates.
PFGE
Digestion of B. hyodysenteriae DNA produced 7–18 and 4–9 fragments for MluI and SalI, respectively. The quality of the gels obtained was high, with clearly defined bands (Fig. 1). All tested isolates yielded a PFGE pattern with at least one of the enzymes, although isolate H19 did not generate any visible pattern when MluI was used. The dendrogram for MluI is shown in Figure 2, with the percentage of similarity ranging from 25 to 100. Reference and type strains B204 and B78T grouped separately for both enzymes.

Fig. 1. PFGE patterns of 24 Spanish B. hyodysenteriae field isolates obtained with (a) MluI and (b) SalI. Lanes 1, 10, 19 and 28 show lambda markers (size range 50–1000 kb). Isolates in lanes 2–9 are 78, 53, 5677/96, 55, 88, 94, 23 and 92. Isolates in lanes 11–18 are 20, 19, 13, 17, 10, 60, 59 and 93. Isolates in lanes 20–27 are 98, 96, 36, H227, H72, H9, H19 and 40.
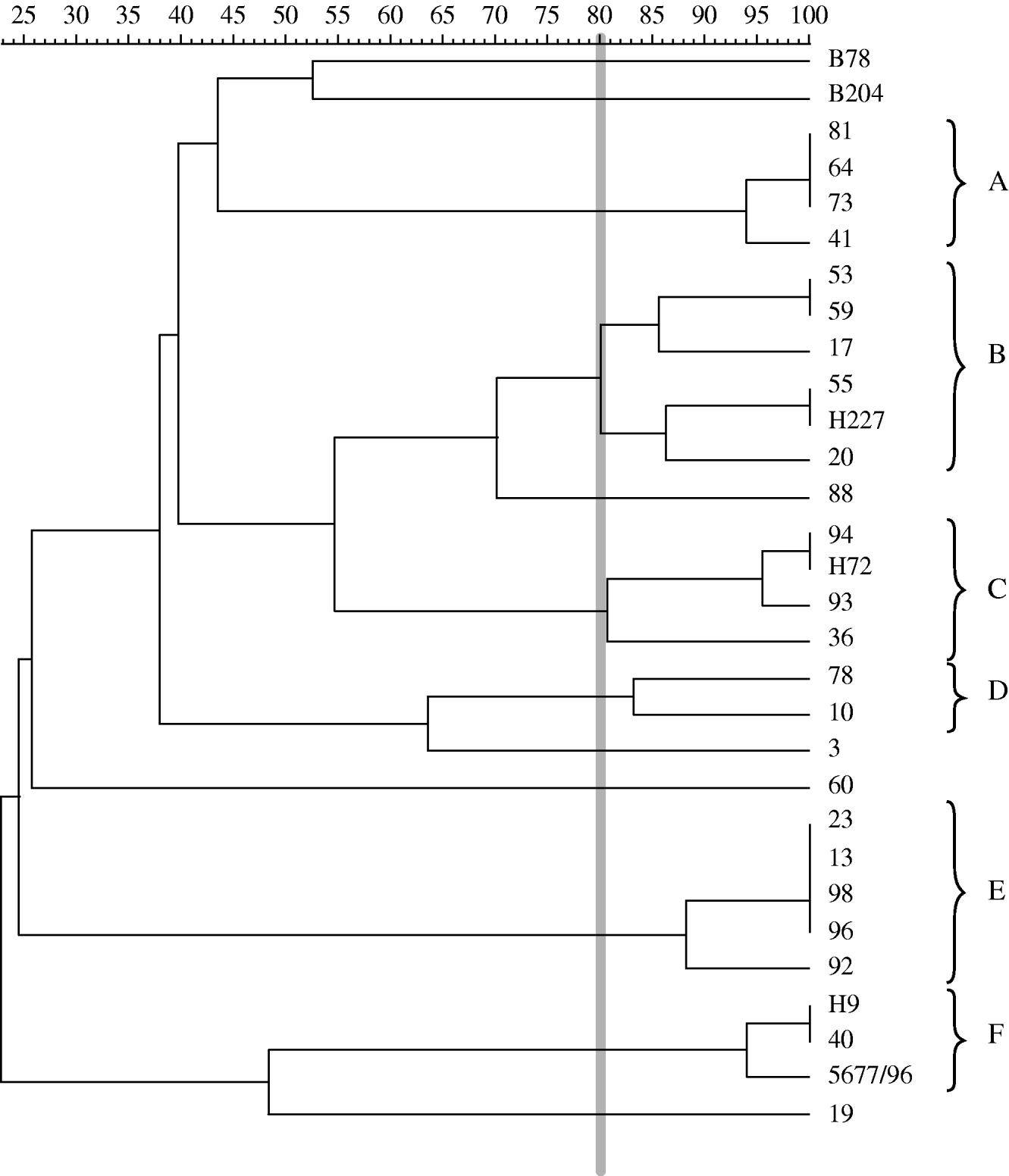
Fig. 2. Dendrogram based on PFGE patterns for MluI clustered by UPGMA strategy and depicting genetic similarity for 31 B. hyodysenteriae isolates, including 28 Spanish field isolates, the reference and type strains of B. hyodysenteriae (B204 and B78T), and one German indole-negative isolate (5677/96). An 80% cut-off value (thick vertical grey line) has been used for establishing groups of related isolates (A–F).
SmpA/smpB analysis
All Spanish B. hyodysenteriae isolates tested were smpA-positive, as revealed by PCR analysis.
DISCUSSION
The combination of strong β-haemolysis and 23S rRNA PCR [Reference Leser13] has been used in the Laboratory of Infectious Diseases in the Veterinary Faculty at the University of León to identify B. hyodysenteriae in spirochaete isolates from swine. In addition, duplex PCR based on the tlyA and 16S rRNA genes [Reference Råsbäck14] confirmed the identification of 70 B. hyodysenteriae isolates which were later studied in detail. This analysis revealed two cultures mixed with B. pilosicoli that were not used in the following procedures. Biochemical tests allowed the further detection of two other mixed Brachyspira spp. cultures that were excluded from the study. These data emphasize the importance of using biochemical tests together with PCR techniques for routine diagnostics, as previously proposed [Reference Fellström20, Reference Råsbäck21].
Several techniques have been applied to characterize B. hyodysenteriae isolates. In the current study we used a combination of RAPD and PFGE for this purpose. This methodology has been previously recommended for other bacteria [Reference Gori22]. It combines the simplicity and promptness of RAPD for establishing groups of closely related isolates with the potency of PFGE as a confirmatory technique for the previously established groups.
In general, RAPD was useful as an initial screening technique for the characterization of B. hyodysenteriae isolates. RAPD patterns were stable and reproducible although the interpretation was sometimes hampered by slight changes in band brightness intensities in the replicates performed for each isolate. Moreover, the use of PFGE allowed us to establish epidemiological connections and to study phylogenetic relationships between isolates. Both restriction enzymes MluI and SalI showed a similar ability to discriminate between isolates and produced analogous clusters when dendrograms were examined. Although it has been reported that PFGE is not always feasible for strongly haemolytic Brachyspira spp. [Reference Råsbäck21], the protocol described in this study, adapted from a protocol for Treponema spp. [Reference Pringle18], produced good quality pulsed-field gels which were suitable for computer processing.
RAPD permitted us to classify 70 Spanish field isolates of B. hyodysenteriae into 28 different patterns. Twenty out of 28 RAPD patterns (71%) belonged to isolates recovered from single herds. However, eight out of the 28 RAPD fingerprints (29%) were shared by isolates from different herds. Isolates with a common RAPD pattern shared geographical origin, e.g. isolates with RAPD patterns 1 and 3 originated from Murcia, and RAPD patterns 8 and 17 originated from Cataluña and Castilla y León, respectively. Other isolates originated from neighbouring regions of Spain: RAPD pattern 5 from Murcia and C. Valenciana; RAPD pattern 20 from Andalucía, Castilla-La Mancha and Extremadura; RAPD pattern 9 from Cataluña and Aragón; RAPD pattern 21 from Castilla-La Mancha and Andalucía (Table 1). As previously proposed [Reference Trott, Oxberry and Hampson7, Reference Fellström10], movements of infected pigs between herds could have facilitated the spread of particular strains within a region. However, where farms are placed in close proximity, infected rodents and drainage effluent might also play a potential role in transmission [Reference Hampson, Atyeo, Combs, Hampson and Stanton2].
A specific PCR for differentiation of smpA/smpB B. hyodysenteriae isolates was designed and performed by Holden et al. [Reference Holden19], who reported a similar distribution of both genes (50% smpA and 50% smpB) in eight B. hyodysenteriae strains from Australia, Canada, UK and USA. Only the smpA gene was detected in the isolates investigated in the current study. According to this result, SmpA, a lipoprotein that has been demonstrated to be a highly immunogenic outer membrane component of B. hyodysenteriae [Reference Sellwood23, Reference Thomas and Sellwood24] should be considered when designing subunit vaccines against SD in Spain. Moreover, this result could have implications in other fields such as serological diagnosis of SD in Spanish farms.
Biochemical characterization confirmed the presence of indole-negative isolates in Spain. Atypical indole-negative B. hyodysenteriae have only been reported previously in Belgium, Germany and Canada [Reference Fellström10, Reference Hommez25]. Further characterization of these isolates with RAPD showed two different banding patterns. One of these patterns was represented by a single isolate, identified as isolate 60, which was recovered in January 2008 in a farm located in the northwest of the country (Castilla y León). The second RAPD pattern was shared by the other eight indole-negative isolates: 40, 43/H170, 44/H137, 46/H181, 45/H138, 50/3140, 51/H3 and H9. These isolates were recovered from seven different farms located in two neighbouring regions in the northeast of Spain, i.e. Cataluña and Aragón, between 2002 and 2008. Surprisingly, this RAPD pattern was also shared by the two indole-negative German isolates, T4 and 5677/96. For further investigation of this relationship, the German isolate 5677/96 together with two Spanish indole-negative isolates, H9 and 40, were analysed by PFGE. The three isolates grouped together markedly separated from other clusters, with a high percentage of similarity (94% for MluI). Moreover, Belgian indole-negative isolates have been previously shown to be indistinguishable from isolate 5677/96 [Reference Fellström10]. The rare occurrence of indole-negative isolates combined with the results of RAPD and PFGE procedures strongly indicates an epidemiological relationship between these isolates, although our epidemiological records do not allow an absolute confirmation of this fact. Nevertheless, the trade of pigs from these countries to Spain supports this possibility, with more than 207 000 animals sold in 2000 and 135 000 in 2001 (source: Spanish Ministry of Environment and Rural and Marine Affairs). Migratory birds may also be considered as a risk for transmission of Brachyspira isolates between countries [Reference Råsbäck21]. The national, seemingly clonal, spread of this indole-negative strain could have been the result of frequent movements and trade of animals in the northeast area of Spain and the presence of this RAPD type (isolates 44/H137 and 46/H181) in one Spanish multiplier herd (no. 19).
The RAPD fingerprints of 20 Spanish B. hyodysenteriae field isolates recovered from Iberian pigs were divided into six different RAPD patterns, designated as 7, 14, 15, 16, 20 and 21. Subsequent analysis by PFGE grouped RAPD type 14 (isolate 53) together with RAPD type 15 (isolate 55) and RAPD type 20 (isolate 64) together with RAPD type 21 (isolate 73). The spread of RAPD type 20, detected in eight Iberian pig units located in the southwest of Spain (Andalucía and Extremadura), is probably a consequence of trade with carriers or diseased pigs. The particular conditions of the Iberian pig market, which is characterized by high demand for a limited number of available pigs and entirely lacking or deficient herd health programmes, could have facilitated this fact.
The key role of carrier swine in within-herd spread of infection [Reference Hampson, Atyeo, Combs, Hampson and Stanton2] was evident in herd no. 13, a semi-intensive Iberian pig unit where SD appeared and was subsequently disseminated to four productive units situated at different locations.
When more than one isolate per herd were analysed by RAPD, we found identical isolates in four herds (nos. 13, 19, 27 and 38). However, slight variations among isolates were recorded in two other herds (nos. 2 and 28). These isolates were subsequently confirmed by PFGE as closely related. Interestingly, vaccination with an inactivated autologous vaccine of B. hyodysenteriae had been implemented in both herds. Herd no. 2 was analysed further, including 12 isolates recovered from March 2007 to February 2008. Vaccination started in May 2007. The RAPD pattern was stable from February 2007 to January 2008, but slight differences were recorded for two isolates recovered in February 2008. This difference was subsequently confirmed by PFGE. Moreover, the antimicrobial susceptibility pattern also changed. MIC values yielded by isolates from March 2007 (3, 2e/H35, 2e/H36, 2e/H37) were compared with those displayed by one isolate from February 2008 (isolate 10). An increase in the sensitivity of two dilution steps (from 16 μg/ml to 4 μg/ml) was observed for the MIC of tiamulin and of three dilution steps (from 128 μg/ml to 16 μg/ml) for the MIC of lincomycin. This new closely related isolate had not been recovered on the farm previously. One explanation for the isolation of new variants of B. hyodysenteriae in the herd may be the introduction of sows from herd no. 1 (Table 1). However, the minor genetic differences recorded could be the result of adaptive advantages, first, by the selective pressure caused by vaccination or second, by the changes in the antibiotic therapy protocols in the farm subsequent to the success of the immunological treatment. A similar result was reported by Atyeo et al. [Reference Atyeo, Oxberry and Hampson8] in Australian herds and the microevolution theory was also proposed as the most plausible explanation.
On the other hand, genetic stability over time for four Spanish B. hyodysenteriae field isolates was also demonstrated. Isolate 50/3140, an indole-negative isolate, was recovered in October 2002 and yielded an identical RAPD pattern to the indole-negative isolate 46/H181, from February 2008. Similar results were obtained for isolates 58/E1090 and 59 recovered in July 2001 and June 2007, respectively, and isolates 62/1502 and 65/H173, recovered from January 2002 and February 2008, respectively. Similarly, using PFGE, isolate 81 from October 2001 was identical to isolate 73, from October 2007. Hence, stability in some Spanish field isolates of B. hyodysenteriae was registered for up to 6 years, in agreement with a previous report on Swedish isolates [Reference Fellström10].
According to Rønne & Szancer [Reference Rønne and Szancer26], B. hyodysenteriae isolates with MICs >4 μg/ml for tiamulin should be considered as resistant isolates. In the current study, 7/11 B. hyodysenteriae isolates selected on the basis of their reduced susceptibility to tiamulin [Reference Hidalgo4] were classified as resistant. The tiamulin-resistant isolates 1/H40, 3, 2e/H35, 2e/H36 and 2e/H37 shared the same RAPD pattern. The RAPD patterns for the other two tiamulin-resistant isolates were unique; thus three different RAPD and PFGE types of tiamulin-resistant B. hyodysenteriae (MIC 32 μg/ml) were confirmed. Valnemulin decreased susceptibility was present in all tested isolates. Subsequent analysis of the geographical distribution of the herds where the resistant isolates had been collected showed that they were from three different and distant areas of the country: Murcia, Castilla y León and Cataluña. The tiamulin-resistant isolates should be considered as a risk to the swine industry.
In conclusion, the results from RAPD and PFGE demonstrated the presence of diverse B. hyodysenteriae field isolates in Spain and allowed the investigation of epidemiological relationships between these isolates. Furthermore, this is the first report of Spanish indole-negative B. hyodysenteriae isolates and the clonal spread of one of these. Moreover, the existence of tiamulin-resistant B. hyodysenteriae isolates, which have emerged independently in Spain, was also demonstrated.
ACKNOWLEDGEMENTS
The authors express their thanks to Marih Jonsson and Gloria Fernández Bayón for excellent technical assistance. Álvaro Hidalgo is supported by a grant from Consejería de Educación of the Junta de Castilla y León and the European Social Fund. This work was funded by the Ministerio de Educación y Ciencia (Spanish Ministry of Education and Science) and co-financed by the European Regional Development Funds (ERDF) as Project AGL2005-01976/GAN (January 2006).
DECLARATION OF INTEREST
None.






