INTRODUCTION
Scrapie is a transmissible spongiform encephalopathy (TSE) affecting sheep and goats. In sheep the ARR haplotype of the prion protein (PrP) gene (PRNP) defined at codons 136, 154 and 171 [Reference Baylis1] confers resistance to classical scrapie. The promotion of this haplotype and reduction of the frequencies of the VRQ and ARQ haplotypes, conferring susceptibility, have been the backbone of the programmes to breed for resistance to classical scrapie in individual and national flocks, in accordance with Commission Regulation (EC) 999/2001 [2]. Similar to the sheep gene the PRNP gene in goats is highly polymorphic, although the evidence for polymorphisms linked to scrapie resistance in goats is less certain [Reference Vaccari3]. Although several studies have shown promising results, the exploitation of goat genetics as a management tool for the control of scrapie has not produced reliable outputs as yet. The substitution of methionine (M) for isoleucine (I) at codon 142 is associated with increased resistance to preclinical and clinical scrapie [Reference Goldmann4, Reference Goldmann5]. Other polymorphisms – histidine (H)→arginine (R) at codon 143, arginine (R)→histidine (H) at codon 154, arginine (R)→glutamine (Q) at codon 211 and glutamine (Q)→lysine (K) at codon 222 – have also been investigated for disease association [Reference Billinis6–Reference Corbière11]. The polymorphisms at codon 146 – N/D/S have shown clear association with disease risk in Cypriot goats: animals carrying serine (S) or aspartic acid (D) at this position have shown resistance to scrapie, while the wild-type allele asparagine (N) appears to be associated with susceptibility to natural scrapie [Reference Papasavva-Stylianou12, Reference Papasavva-Stylianou13]. Some experimental studies recently published support a potentially high level of resistance for goats carrying the S146 and K222 alleles [Reference Acutis14–Reference Lacroux16].
Scrapie is highly prevalent in Cyprus causing severe losses to goat production. A previous study of 250 Cypriot goats, 164 of which were histologically positive for scrapie, found that polymorphisms at codon 146 were associated with resistance or susceptibility to natural disease [Reference Papasavva-Stylianou12]. Polymorphisms S146 or D146 were found only in healthy controls and scrapie-negative goats. In a larger study of 717 Cypriot goats, including 218 scrapie-positive animals, amino acids N and R at codons 146 and 154, respectively, were associated with susceptibility to goat scrapie. Animals heterozygous for S146 and D146 were significantly under-represented in scrapie-positive animals and there were no positive animals among those homozygous for these amino acids at codon 146 [Reference Papasavva-Stylianou13].
The scientific validity of these results was evaluated by the BIOHAZ panel of the European Food Safety Authority who acknowledged them as additional proof of a potentially lower susceptibility to classical scrapie in goats that were H154, D146 and S146 PRNP allele carriers compared to the wild type (NN146) [17–19]. In order to investigate the possibility that putatively resistant animals were subclinically infected and to further elucidate the resistance to scrapie in natural conditions of animals carrying D146 or S146, a systematic assessment of the detectability of PrPSc in the central nervous system and peripheral tissue of clinically healthy and scrapie-affected animals from long-term infected herds was conducted; the results are presented in this paper.
MATERIALS
The Cypriot Veterinary Services identified candidate scrapie-affected herds, with a minimum herd size of 200 goats aged >12 months, which had the largest proportion of positive cases in animals submitted for TSE testing in the previous 2 years. The owners of eligible herds were approached, and four agreed to cull their entire herds and receive compensation. The cull took place in 2009 at two culling centres managed by the Cypriot Veterinary Services. This sample size provided 90% power to detect an infection prevalence difference of 20% between NN146 and non-NN146 genotypes assuming a 5% frequency of non-NN146 genotypes and a 25% prevalence of scrapie infection in the culled population.
The animals were culled and samples taken from key anatomical sites associated with classical scrapie pathogenesis. Whole brain, thoracic spinal cord (T10–12), distal ileum and lateral retropharyngeal lymph nodes (LRPLN) were collected from each animal aged >12 months, with half being retained frozen at −20 °C and the other half fixed in 10% formol saline. The fixed brain samples were blocked to represent the brainstem (obex), cerebellum, midbrain, basal ganglia and frontal cortex. All fixed tissues were processed using standard histological methods, and embedded in wax. For immunohistochemistry (IHC), 5-μm sections were immunolabelled using a standardized protocol [Reference Simmons20], and primary antibody MAb 2G11 (AbD Serotec, USA). This antibody, raised against ovine PrP peptide sequence 146-R154R171-182, and specifically recognizing the R151–R159 sequence, has been shown to label experimental positive cases in N146, S146 and D146 allotype carriers [19].
The infection status of each animal was assessed by testing fresh brainstem (obex) for the presence of PrPSc using the Bio-Rad TeSeE® ELISA Detection and Purification Test kit (Bio-Rad Laboratories Ltd, USA), and screening of the fixed obex and LRPLN by IHC. An animal was considered infected if it was positive in any of the tests conducted.
DNA was extracted from goat brain material using the DNeasy Blood & Tissue kit (Qiagen, Germany) according to the manufacturer's instructions. In order to inactivate potential TSE infectivity from the extracted DNA, a phenol inactivation step was carried out followed by its removal with chloroform. PCR amplification was performed with Roche Genotyper master mix for Light Cycler 480 and a primer probe mix which discriminates codons 146 and 154 of the caprine PRP gene (TipMolBiol, Germany). Primers used for DNA sequencing were cPRNP_F (gtggctacatgctgggaa) and cPRNP A (ggagaaaagaggatcacacttgcc).
Age, sex, herd and genotype were collated for all the animals from the culled herds and imported into an ad hoc database together with test results.
A Bayesian multivariate logistic regression model was fitted using scrapie status as the dependent variable and genotype (NN146, all other genotypes/unknown), age (quartiles), herd and sex as explanatory variables. An extra covariate was introduced in the model in order to verify that the impact of the sensitivity of the diagnostic tests and the temporal pattern of the prevalence of infection in each herd had no influence over the final estimates of the risk. In particular, this approach allowed inclusion of the sensitivity of each test [Reference Branscum, Gardner and Johnson21], as follows. Let the sensitivity of the rapid test, IHC applied to the obex, and IHC applied to lymphoreticular tissue (LRT) be denoted by θ
RT, θ
obex, and θ
LRT, respectively. The test results for the ith animal are denoted by
![]() $\lambda _i (j,k,l)$
, where j, k, l represent the test results for the rapid test, IHC applied to the obex and IHC applied to the LRT; j = 0 if the result for the rapid test is negative and j = 1 if the rapid test result is positive, with the same applying to k and l for the obex and LRT results, respectively. Then the probability that an animal is negative for all three tests, is given by
$\lambda _i (j,k,l)$
, where j, k, l represent the test results for the rapid test, IHC applied to the obex and IHC applied to the LRT; j = 0 if the result for the rapid test is negative and j = 1 if the rapid test result is positive, with the same applying to k and l for the obex and LRT results, respectively. Then the probability that an animal is negative for all three tests, is given by
where
![]() $\pi _i $
denotes the probability that an animal is infected, which is the product of the relative risk by genotype, the flock-level prevalence and the relative risk of being positive according to age group. The probability that an animal is positive for at least one of the tests is given by
$\pi _i $
denotes the probability that an animal is infected, which is the product of the relative risk by genotype, the flock-level prevalence and the relative risk of being positive according to age group. The probability that an animal is positive for at least one of the tests is given by
 $${\eqalign{\lambda _{_i} (\,j,k,l) &= \pi _i (1 - \theta _{\rm RT} )^{1 - j} \theta _{\rm RT}^j (1 - \theta _{\rm obex} )^{1 - k} \theta _{\rm obex}^k \cr &\quad\times(1 - \theta _{\rm LRT} )^{1 - l} \theta _{\rm LRT}^l. }}$
$${\eqalign{\lambda _{_i} (\,j,k,l) &= \pi _i (1 - \theta _{\rm RT} )^{1 - j} \theta _{\rm RT}^j (1 - \theta _{\rm obex} )^{1 - k} \theta _{\rm obex}^k \cr &\quad\times(1 - \theta _{\rm LRT} )^{1 - l} \theta _{\rm LRT}^l. }}$
The resulting data for each animal then follow a multinomial distribution with eight possible outcomes (each of three test results being positive or negative), the likelihood of each outcome being determined by λ i (i, j, k). Goats for which test results from the three target areas were not available were removed from the analysis, leaving 1060 animals in the final dataset. The relative risk by age was estimated using quartiles. Herd prevalence was estimated for each herd but Deviance Information Criterion (DIC) [Reference Spiegelhalter22] was used to determine whether there was any significant influence of the change in the prevalence of infection over time in each herd (estimated for each age quartile) on the model fit.
Estimates of the test sensitivities, relative risk by genotype and age, and the prevalence of infection in each herd were estimated using 15000 iterations following a burn-in of 5000. All analyses were conducted using WinBUGS v. 1.4 (http://www2.mrc-bsu.cam.ac.uk/bugs/winbugs/contents.shtml).
RESULTS
Four herds were culled within the scope of the study. Three of them fulfilled the selection criteria but the fourth had only 37 goats at the time of the cull. The proportion of positive animals from those submitted for TSE testing was among the highest of all known scrapie-affected herds, according to the surveillance data provided by the Cypriot Veterinary Services (Table 1). In all other aspects the culled herds were representative of the goat herds in the country: they had been established for at least 15 years before being culled and were managed under extensive production systems using natural mating.
Table 1. Surveillance data results for the four herds culled for the period 2006–2009

* Herd size according to census data extracted in May 2009.
A total of 1075 goats aged >12 months were culled with the following distribution (herds are identified by capital letters): 257 (A), 37 (B), 394 (C) and 387 (D). Overall the average age of the culled population was 46·7 months (3·9 years) with the youngest animal culled aged 14 months (1·16 years) and the oldest 135 months (11·2 years).
The majority of animals in the four herds were NN146 (Table 2), ranging from 65·4% to 84%. There was variation in the frequency of putative semi-resistant genotypes at herd level (ND146 and NS146): from 2·5% to 23·7% for ND146 frequency and from 0% to 13·2% for NS146 frequency. The putatively resistant genotype DS146 was not present in three herds and only accounted for 1·2% in one herd. The other putatively resistant genotypes (DD146 and SS146) appeared at a very low proportion with one single SS146 animal in herd D and three having DD146, two in herd A and one in herd C. There was no significant association between age group (⩽2 years, >2–4 years, >4 years) and genotype as a binary variable (NN146, all other 146 genotypes) overall for the whole culled population (χ 2: P = 0·6) and individually for three herds (χ 2: P = 0·2, P = 0·5, P = 0·4) for herds A, B and C, respectively. Only in herd D were NN146 goats older than non-NN146 goats (P = 0·05) Table 3 shows the distribution of goats in the four culled herds by age and genotype categories. By alleles, D146 and S146 had similar frequencies: 4·9% and 5·9%, respectively, whereas N146 was present in 89·1% of all possible alleles. There was a great variability of allele frequencies between herds: allele D146 frequency ranged between 1·5% and 13·2%, allele S146 frequency between 0% and 6·6% and allele N146 frequency between 81·7% and 91·8%.
Table 2. Codon 146 genotype distribution of the four culled herds with percentages in parentheses and total

Table 3. Number of goats with NN and non-NN genotypes (% of the total) by age group in the study population

There were 234 positive cases detected by either the rapid test or IHC in any of the tissues selected. The overall proportion of positive cases in the four culled herds was 21·7%. There were 73, 21, 80 and 60 positive cases in herds A, B, C and D, respectively, which represents within-herd prevalences of 28·4%, 56·7%, 20·3% and 15·5%. All positive cases except one, which was of undeterminable genotype, were of the NN146 genotype and had the following test profile: positive by rapid test and by IHC for both the obex and LRT. One hundred and seven positive cases (45·3%) were positive by rapid test and IHC for both the obex and LRT. Only one case (NN146 genotype) was positive by the rapid test and negative by IHC, whereas 75 (32%) cases were only positive by IHC in LRT. Six (2·5%) cases were negative by IHC in LRT and positive by the rapid test and IHC in the obex. All 30 cases positive by IHC for both the obex and LRT and negative by the rapid test were NN146 genotype (Table 4).
Table 4. Summary of the test results of the 1075 culled animals by test
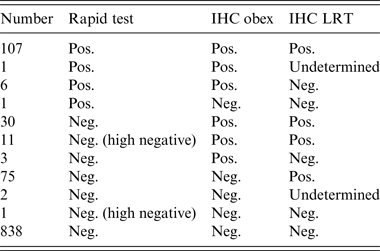
IHC, Immunohistochemistry; LRT, lymphoreticular tissue (lateral retropharyngeal lymph node and/or distal ileum).
The final logistic regression model included genotype, herd prevalence, age and sensitivity of the tests. The odds of scrapie infection occurring in NN146 goats was 101 [95% credible interval (CrI) 19–2938] times higher than for non-NN146 or unknown genotypes (one case of undeterminable genotype included in this group, as described above). There were significant differences in within-herd prevalences of infection but there was no significant improvement in model fit by including an effect of prevalence over time (P = 0·45). Positive cases in goats aged 27–42 months were three times more likely than cases in goats younger than 26 months (P < 0·001), adjusted for genotype and herd. The odds of being scrapie positive were twice as high in goats aged >42 months compared to goats aged <26 months (P < 0·05) adjusted for genotype. IHC applied to LRT produced the highest sensitivity (94%, 95% CrI 90–97), followed by IHC for the obex (64%, 95% CrI 58–71) and the rapid test (50%, 95% CrI 44–57). Final estimates of the significant parameters, i.e. median of the estimated odd ratios and 95% CrIs, are displayed in Table 5. DIC indicated that there were no significant differences between the estimated relative risk for ND146, NS146 and DS146 genotypes; hence these were grouped together for the relative risk estimation.
Table 5. Results of the logistic regression model showing median of the estimated odd ratios and 95% credible intervals (CrI)

When assessing the effect of imperfect test sensitivity by the use of a Bayesian model, there was no evidence in the data that a significant number of positive animals were missed by imperfect diagnostic tests since tests were applied in parallel, with the model estimating a high sensitivity for the LRT IHC test.
DISCUSSION
This study attempted to estimate the level of resistance to scrapie in goats, with different codon-146 genotypes, born and reared in a scrapie-infected environment, by investigating the distribution of PrPSc in the central nervous system and in peripheral tissues. The results are consistent with those reported in the previous studies conducted in Cyprus [Reference Papasavva-Stylianou12, Reference Papasavva-Stylianou13] and confirm that Cypriot goats with amino acids serine (S) or aspartic acid (D) at codon 146 show an association with resistance to natural scrapie. Although degrees of resistance could not be determined in this study, the failure to detect any positive goats with non-NN genotypes allows speculation that S146 or D146 heterozygosity confers a certain level of resistance compared to NN146 goats under natural conditions. Intracerebral and oral challenge experiments, conducted in parallel with this study, which are still ongoing will elucidate degrees of codon 146-linked disease risk, if they occur. However, a recent oral scrapie challenge experiment showed that S146 heterozygotes remained clinically normal and negative by rectal biopsy at significantly longer post-inoculation times than the N146 homozygous goats [Reference White15].
The panel of tissues tested represented a pragmatic approach to the major systems known to be involved in TSE pathogenesis in small ruminants. This selection was based on the knowledge available at the time regarding the full PrPSc distribution in Cypriot goat scrapie (M. Groschup, personal communication) and from caprine data previously presented by Gonzalez et al. [Reference Gonzalez23].
The concordance of the rapid test and IHC test results in the obex revealed a high agreement (kappa coefficient 0·81) with only 45 discordant results (44 positive by IHC and negative by rapid test), but not as high as the 0·98 reported in similar anatomical area by [Reference Corbiere24]. However, the concordance level was moderate when considering IHC in both the obex and LRT (kappa agreement 0·59) due to the 75 animals that were only positive by IHC in LRT. When considering only positive samples, the low sensitivity shown by the rapid test raises questions about its efficiency as a primary screening test in surveillance of scrapie in goats. The fact that a large proportion of positive animals was not detected by the rapid test has been highlighted in a previous study where a similar testing regimen was conducted after the cull of scrapie-affected herds in the UK [Reference Gonzalez23]. The unreliability of the notification of clinical suspects by farmers dictates that active surveillance regimens using currently available TSE screening tests are the main detection tool for identifying newly infected herds. However, as shown in a recent study, the performance of TSE detection in goats varies according to age and targeted tissue [Reference Corbiere24]. Tests currently available for use for TSE surveillance in small ruminants at the EU level were evaluated and approved on behalf of the European Commission using only ovine samples [25]. These data suggest that it would be prudent to review test performance specifically for goats.
Data produced by this study provide evidence that the selection for breeding of D146 or S146 goats can be applied to reduce the incidence of classical scrapie with the ultimate objective of eliminating the disease from the Cypriot goat population. The frequency of these alleles in the goat population will determine the availability of breeding goats with which to disseminate putative-resistant alleles. In our study population, the frequencies of the D146 and S146 alleles were 4·9% and 5·9%, respectively. These frequencies although still low are higher than the study population described by Papasavva-Stylianou et al. [Reference Papasavva-Stylianou12] (comprising 250 goats with frequencies of 4·2% and 4·4% for D146 and S146, respectively), and of a similar study by the same authors among 717 goats in 2011 with frequencies of 4·2% and 4·7%, respectively [Reference Papasavva-Stylianou13]. These differences could be due to sampling variability or to the fact that our study did not include controls from non-scrapie positive herds, i.e. the sampling populations were different. In any case the low frequency of the D146 and S146 alleles in the Cypriot goat population does not preclude the feasibility of a breeding programme based on the promotion of either or both alleles.
CONCLUSION
In a heavily infected goat population, susceptibility is linked to the NN146 genotype of the PrP gene. Being NN146 is a statistically significant risk factor for the presence of detectable infection. In our study no positive cases were detected among all other available genotypes. The epidemiological evidence for the resistance to natural scrapie at population level associated with allelic variants of the goat PrP gene at position 146 confirms the results of previous case-control studies conducted in the same population and paves the way for experimental studies that might confirm the modulation of susceptibility to scrapie by polymorphisms at codon 146. Although represented at a low frequency at the time of conducting this study, the presence of these putatively resistant alleles in the Cypriot goat population provides a potential tool to reduce/eradicate scrapie provided that nationwide breeding programmes are implemented and maintained over time.
ACKNOWLEDGEMENTS
This study has been partly funded by the Cypriot Government and the European Commission through the EU Reference Laboratory for TSEs. The authors are grateful to the owners of the participating herds, staff of the District Veterinary Offices, the Histopathology and TSE laboratories of the Cypriot Veterinary Services, staff of the Pathology Department, AHVLA Weybridge for logistical and technical support, and to Saira Cawthraw, Melanie Chaplin and Linda Davies of the TSE Department, AHVLA Weybridge.
DECLARATION OF INTEREST
None.








