INTRODUCTION
Cholera remains an important public health problem in Africa and Asia. Several countries in both these continents are known to be endemic for cholera. At the beginning of the seventh pandemic during the 1960s, cholera had a strong presence in Asian countries. In the early 1970s, the pandemic spread to sub-Saharan Africa and caused explosive outbreaks with a high case fatality, mainly because of a lack of background immunity in the population. During this pandemic, cholera spread along the coast and into the interior through waterways and further spread into the interior of the Sahelian states by land travel fostered by nomadic tribes [Reference Kaper1, Reference Faruque, Albert and Mekalanos2]. Based on cholera reports covering a period of more than 10 years (1995–2005), it is evident that about 88% of cholera originated in sub-Saharan Africa and the rest from South East Asia [Reference Griffith3], and a 5·4% case-fatality rate (CFR) was reported between 2000 and 2005 in the eastern provinces of Congo [Reference Bompangue4]. Thirty-one (78%) of the 40 countries which reported cholera cases to WHO in 2005 were from sub-Saharan Africa with an incidence rate of 166 cases/million population and recently, cholera outbreaks have spread to newer African countries like Namibia [Reference Smith5].
The cholera statistics released recently by WHO paint a gloomy picture with both the number of cases and the number of countries reporting cases to WHO increasing during 2008, reaching the level of the late 1990s [6]. The overall number of notified cases of cholera to WHO during the most recent 5-year period (2004–2008) showed a 24% increase in the number of cases compared to cases between 2000 and 2004 [6]. Globally, there was a 27·5% increase in the number of deaths, representing an overall CFR of 2·7%. Of the total number of deaths, 98% were reported from the African continent. In vulnerable groups living in areas at high risk for cholera, the CFR reached as high as 50% [6]. During 2004, major outbreaks of cholera occurred in Cameroon, Chad, Guinea, Mali, Niger, Senegal and Zambia [7]. Cholera surveillance in Beira, Mozambique, in 2004 [Reference Ansaruzzaman8] revealed the presence of a distinct El Tor strain that displayed most of the typical traits of the El Tor biotype but resident CTX prophage in the strain was of the classical type (CTXclass φ). Since Zambia shares a common border with Mozambique, cholera surveillance in Zambia between 1996 and 2004 [Reference Mwansa9] showed that Zambian V. cholerae O1 El Tor strains are not hybrids based on the repressor gene (rstR) of the CTX prophage and showed typical seventh pandemic strains of the El Tor biotype [Reference Finkelstein10].
The pathogenesis of cholera is mainly related to the production of cholera toxin (CT) encoded by the ctxAB gene, which is located on the CTX prophage integrated on the V. cholerae chromosome which has two epitypes or immunological forms: CT1 displayed by classical biotype strains and by US Gulf Coast strains, and CT2 demonstrated by the El Tor biotype and O139 strains [Reference Finkelstein10]. These two toxins differ in the CT B subunit by 2/124 amino acids, and a polymerase chain reaction (PCR)-based method to discriminate classical-type ctxB and El Tor-type ctxB has recently been described [Reference Morita11]. Another classification recognizes three genotypes based on the ctxB gene: genotype 1 is observed in classical strains and US Gulf Coast strains, genotype 2 is found in El Tor biotype strains from Australia, and genotype 3 is observed in El Tor biotype strains from the seventh pandemic and the Latin American epidemic [Reference Olsvik12]. The DNA sequences of the ctxB gene of Zambian V. cholerae have been deposited in GenBank under accession numbers EU932878-EU932884.
Recently, genetic hybrids of V. cholerae O1 strains possessing both the properties of classical and El Tor biotypes were found in Bangladesh [Reference Nair13, Reference Nusrin14] and studies also showed that the CT genotype of the El Tor strains currently associated with cholera in Bangladesh have shifted from genotype 3 to genotype 1. It has been shown that the seventh pandemic prototype El Tor strains had been replaced by El Tor variant strains in Kolkata and Bangladesh from 1995 and 2002, respectively [Reference Nair15, Reference Raychoudhuri16]. The genetic analysis results showed that these altered strains vary by CTX prophage type and presence of the RS1 element, although the exact array of the CTXΦ and RS1 element and their location on the genome of these strains remain to be clarified. Comparative genomic analysis using a DNA microarray showed differences in gene content between the sixth (classical biotype) and current seventh (El Tor biotype) pandemic strains of V. cholerae O1 and identified two genomic regions designated as the Vibrio seventh pandemic island-I (VSP-I) and VSP-II that are unique to seventh pandemic El Tor strains [Reference Dziejman17].
In view of rapidly growing genetic diversity in toxigenic V. cholerae strains with epidemic potential, we conducted detailed genetic analysis on the Zambian V. cholerae O1 strains isolated between 1996 and 2004 to better understand the genetic organization of Zambian strains.
MATERIALS AND METHODS
Bacterial strains
Sixty-nine strains of V. cholerae O1 isolated between 1996 and 2004 in Zambia were included in this study. Of the 69 strains, 27 were isolated in 1996/1997 and 42 strains were isolated in 2003/2004. Three V. cholerae O1 strains isolated between 1991 and 1994, designated as Matlab type 1 (MJ-1236), type II (MG-116226) and type III (MG-116955) [Reference Nusrin14] and two more strains, one (B-33) isolated in Beira, Mozambique in 2004 [Reference Mwansa9], one V. cholerae O1 isolated in Dhaka, Bangladesh in 2001 [Reference Dziejman17] were also included in this study for comparison. We used V. cholerae O1 strain N16961 of El Tor biotype and O395 classical biotype as reference strains. All strains were grown in Luria Bertani broth (LB) and stored frozen (−80°C) in LB broth containing 25% (v/v) glycerol.
Genomic DNA isolation
For extraction of genomic DNA, cells were harvested from 3 ml of overnight culture in LB broth (Miller). The harvested cells were subjected to alkaline lysis by 10% SDS in the presence of TE buffer (10 mm Tris–HCl, 1 mm EDTA; pH 8·0). The cells were then treated with freshly prepared Proteinase K (final concentration 100 μg/ml in 0·5% SDS) and incubated at 37°C for 1 h. After incubation, 1·0% CTAB/NaCl (cetyl trimethyl ammonium bromide in 0·7 m NaCl) was added followed by incubation for 10 min at 65°C. RNA was removed by treating with RNaseA (Sigma, USA, code no. R6513) at 37°C for 1 h. This was followed by phenol chloroform extraction and precipitation of the nucleic acid in the presence of isopropanol [Reference Chowdhury18]. Excess salt was removed by 70% alcohol wash and the nucleic acid was air-dried, resuspended in sterile TE buffer and the purity assayed using a spectrophotometer (Gene Quant, UK) that calculates the ratio of optical densities at 260 nm and 280 nm. The DNA was stored at −20°C for subsequent PCR analysis.
Mismatch amplification mutation PCR assay (MAMA)–PCR analysis
PCR assays were performed to test for the presence of ctxB genes specific for classical and El Tor biotypes. A conserved forward primer (Fw-con, 5′-actatcttcagcatatgcacatgg-3′) and two allele-specific polymorphism detection primers, Rv-cla (5′-cctggtacttctacttgaaacg-3′) and Rv-elt (5′-cctggtacttctacttgaaaca-3′) [Reference Morita19] were used in this study. PCR conditions were as follows: initial denaturation at 96°C for 2 min, 25 cycles of denaturation at 96°C for 10 s, annealing at 50°C for 10 s, and extension at 72°C for 30 s, with a final extension at 72°C for 2 min. V. cholerae O1 isolates, O395 classical and N16961 El Tor were used as standard reference strains.
Multilocus virulence gene profiling
We used a multilocus virulence gene profiling strategy that scanned for nine virulence-associated genes and/or gene clusters in the genome of 68 representative Zambian V. cholerae O1 isolates by year (1996–2004) using 33 sets of PCR primers and conditions as described previously [Reference Nusrin14, Reference Chow20–Reference Keasler and Hall23]. PCR was performed in 20 μl reaction mixture as follows: initial denaturation step at 96°C for 1 min followed by 30 cycles of denaturation at 94°C for 30 s, primer annealing at 45–58°C for 30 s, 1–4 min of primer extension at 72°C and 7 min of final extension at 72°C for one cycle. The PCR products were analysed by electrophoresis in 1% agarose gels, stained with ethidium bromide, visualized under UV light and recorded by a gel documentation system (Gel Doc™ 2000, Bio-Rad, USA). The PCR products were sized with standard molecular-weight markers and documented.
Nucleotide sequence of ctxB subunit
To determine the nucleotide sequence of the ctxB subunit of CT, PCR amplification of ctxB genes of eight representative strains of V. cholerae O1 based on VSP-II pattern resulting from MAMA–PCR, that were isolated in Zambia during 1996/1997 and 2003/2004, was performed in a 25 μl reaction mixture in an automated Peltier thermal cycler (PTC-200, MJ Research, USA). PCR primers and conditions were as previously described [Reference Mitra24]. PCR products were purified with a Microcon centrifugal filter device (Millipore Corporation, USA) and sequenced using an ABI PRISM BigDye Terminator Cycle Sequencing Reaction kit (PerkinElmer Applied Biosystems, USA) on an ABI PRISM 310 genetic analyser (Applied Biosystems).
DNA sequence analysis
The chromatogram sequencing files were inspected using Chromas 2.23 (Technelysium, Australia). Nucleotide sequences of the test isolates were compared with the corresponding sequences of the N16961 El Tor reference strain (NC_002505), the 569B classical reference strain (U25679), retrieved from GenBank using Basic Local Alignment Search Tool (BLAST) [Reference Altschul25]. Multiple sequence alignments were developed using CLUSTALX 1.81.13, and DNA sequences were translated using GeneDoc version 2.6.002 alignment editor (http://www.psc.edu/biomed/genedoc).
Pulsed-field gel electrophoresis (PFGE)
Intact agarose-embedded chromosomal DNA from V. cholerae isolates was prepared and PFGE was performed using a contour-clamped homogeneous electric field apparatus (CHEF Mapper, Bio-Rad) following the standardized PFGE protocol [Reference Cooper26]. Genomic DNA was digested with NotI enzyme (10 U/μl, Invitrogen, USA). The restriction fragments were separated in 1% SeaKem Gold (USA) agarose in 0·5× Tris-borate-EDTA buffer. V. cholerae N16961 (El Tor) and 569B (classical) were used as reference strains to which the test strains were compared. Salmonella enterica serotype Braenderup strain H9812 was used as the molecular size marker. Following electrophoresis, the gels were stained in ethidium bromide solution (0·5 μg/ml) for 20–30 min and destained with reagent grade water. Images were captured using Gel Doc 2000 and Gel Doc XR systems (Bio-Rad). The test fingerprint image was normalized with Quantity One software (Bio-Rad), and the molecular weights of DNA fragments were determined. The degree of genetic diversity between the new variants and the classical and El Tor strains was determined by using Diversity Database software (version 2.2; Bio-Rad) and BioNumeric software (Applied Maths, Belgium). The similarity between the strains was determined using the Dice coefficient, and cluster analysis was performed using the unweighted pair-group method using average linkages (UPGMA).
RESULTS AND DISCUSSION
All 69 V. cholerae O1 strains were serologically confirmed as serogroup O1 and Ogawa serotype. Genetic screening by PCR revealed that all strains except two had important epidemic virulence markers namely ctxA, rstR2 and tcpA El Tor. One strain isolated in 1996 lacked ctxA.
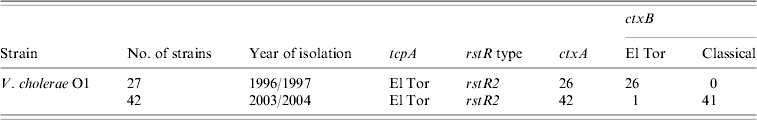
The ctxB genes of the 69 V. cholerae O1 strains isolated between 1996 and 2004 were examined by the primers specific for classical and El Tor biotypes. All strains, except one (ctxA negative) isolated in 1996 were positive for the ctxB El Tor gene while those isolated in 2003 and 2004 were positive for ctxB classical gene, except for one (Table 1) which was positive for ctxB El Tor gene isolated in 2003.
Table 1. Results of ctxB of Vibrio cholerae O1 strains from Zambia
All 69 V. cholerae O1 strains were selected for multilocus virulence gene profiling that involved examining for nine virulence-associated genes and/or gene clusters. All strains showed the presence of all genes comprising VSP-I, MSHA, and RTX gene clusters (Table 2). All the strains isolated in 1996 and 1997 showed the presence of all ORFs in the VSP-II gene clusters whereas all strains isolated in 2003 and 2004 showed the presence of all ORFs in the VSP-II region except the two ORFs, i.e. VC0493 and VC0498 (Table 2). Two individual loci, hlyA and pileE, were also present in all the test strains and the classical and El Tor reference strains examined (Table 2). In the VPI-I gene cluster the major virulence-associated genes; toxT, tcpA and acfB were present in all strains. All the strains were positive for rstA, orfU, zot and ctxAB genes (Table 2). The recognized allele, rstR2 of the repressor gene rstR was present in all the strains (Table 1). Apart from this, two other genes, namely tlc, which is present adjacent to the CTX prophage and integron (intl4), were present in all the strains (Table 2). The overall results of the virulence genetic screening indicated that all the V. cholerae O1 strains isolated in 1996–1997 were similar to the reference El Tor strain N16961. The strains isolated in 2003–2004 carried rstR of El Tor type (rstR ET) but produced CTXB of classical type. Interestingly, these strains isolated during 2003–2004 in Zambia and lacking two ORFs, i.e. VC0493 and VC0498 in the VSP-II region, completely replaced the seventh pandemic progenitor strains and produced outbreaks with full virulence fitness. Nusrin et al. reported that strains isolated between 1991 and 2003 in Peru were negative for ORFs VC0512–VC0515 in the VSP-II gene cluster and concluded that the Peruvian strains had independently evolved new variants [Reference Nusrin27]. It has also been reported that V. cholerae O1 El Tor strain CIRS101 [Reference Nair15] was isolated in 2002 in Bangladesh, another variant of VSP-II, with deletion of ORFs VC0495–VC0512 [Reference Taviani28]. The El Tor strains isolated between 2003 and 2004 in Zambia produced classical ctxB, a new El Tor variant with a deletion of two different ORFs, VC0493 and VC0498 of the VSP-II gene cluster that was not reported earlier.
Table 2. Results of the genetic screen used for identification of nine virulence regions of Vibrio cholerae O1 strains isolated in Zambia between 1996 and 2004
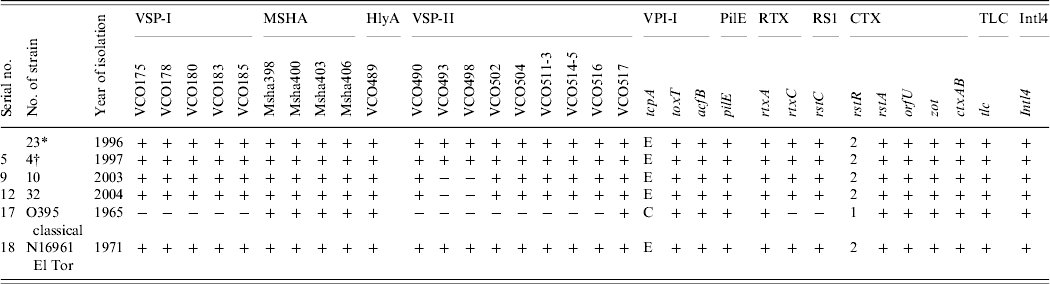
* One strain was negative for ctxAB.
† One strain isolated in 2003 showed an identical pattern of the VSP-II region-like strains isolated in 1997.
VPI-I is a large DNA region that is composed of two gene clusters, i.e. toxin co-regulated pilus (TCP) and accessory colonization factor (ACF) [Reference Herrington29, Reference Kovach, Shaffer and Peterson30]. The tcpA gene of the TCP cluster has alleles specific for classical and El Tor biotypes of O1. In this study, tcpA gene analysis showed that all the tested strains are of El Tor type (Table 2). Another important gene of this cluster is toxT, which encodes a second positive regulatory protein that directly activates a number of virulence genes [Reference DiRita31]. This gene was present in all the tested strains indicating the presence of an active virulence-gene regulatory system. The genes of the MSHA cluster are expressed by strains of the El Tor biotype, but are only rarely expressed by strains of the classical biotype [Reference Jonson, Holmgren and Svennerholm32]. However, all strains in this study were positive for the MSHA gene cluster, including the classical reference strain. The hlyA gene is reported to be present in all classical, El Tor and non-O1 strains of V. cholerae [Reference Brown and Manning33]. PCR study showed that all the tested strains were positive for this gene. The presence of pilE further indicated the pathogenic potential of these strains. PCR assay of the CTX genetic elements indicated the presence of an intact core region of the CTX prophage genome. It has been shown recently that RS1 is a self-transducing phage, and that the rstC gene acts as an anti-repressor in the phage replication process [Reference Davis and Waldor34]. This gene is unique to the strains of the El Tor biotype and is absent in classical strains. All the strains contain this rstC, which grouped them as El Tor biotype, which are different from the Mozambique type which is negative for rstC.
The VSP-I gene cluster encompasses a 16 kb region from VC0175 to VC0185, and most of the genes encode hypothetical or conserved proteins with no known function. On the other hand, the VSP-II region is a ~27 kb region that encompasses VC0490–VC0516 [Reference Rivera22]. As these two clusters are unique to the El Tor strains of the seventh pandemic, we checked for the presence of VSP-I and VSP-II in the genome of the Zambian strains. All strains examined were positive for VSP-I and VSP-II, indicating the seventh-pandemic El Tor ancestry of these strains, PCR assays with primer pair VC0517F–VC0517R showed that VC0517 was present in all Zambian V. cholerae O1 including the classical and El Tor reference strains. The only variation was seen in the strains isolated in 2003 and 2004 that lacked the two ORFs, VC0493 and VC0498 in the VSP-II region. How these two ORFs were deleted and what functional impact it has at this point is still unknown and requires further characterization.
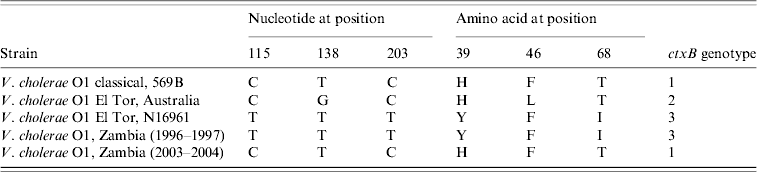
Nucleotide sequence analysis of the ctxB genes of V. cholerae O1 strains isolated between 1996 and 2004 using specific primers [Reference Mitra24] was performed and showed that the strains isolated in 1996 and 1997 had tyrosine at position 39, phenylalanine at position 46 and isoleucine at position 68 which is identical to the El Tor reference strain N16961, whereas the strains isolated in 2003 and 2004 had histidine at position 39, phenylalanine at position 46 and threonine at position 68, which is identical to the classical reference strain 569B and indicated its relationship with classical biotype.
On the basis of DNA and protein sequencing, heterogeneity within the B subunit was first reported in the early 1990s [Reference Kaper1, Reference Brickman35], and since then ctxB typing has been used as a tool for differentiating V. cholerae strains. To date, three ctxB genotypes of V. cholerae O1 strains of both biotypes and serotypes have been identified globally [Reference Nair13, Reference Mekalanos36]. Based on the amino-acid residue substitution at the three positions 39, 46 and 68, all classical and El Tor strains from the Gulf Coast of the USA are categorized as ctxB of genotype 1, El Tor strains associated with the Australian environmental reservoir are genotype 2, and El Tor strains of the seventh pandemic and the recent Latin American epidemic are classified as ctxB genotype 3. In this study, all strains isolated in 1996–1997 belonged to genotype 3 and all strains, except one, isolated in 2003–2004 grouped into genotype 1 (Table 3).
Table 3. Genotypes of Vibrio cholerae O1 strains of Zambia based on the DNA sequence of the cholera toxin B subunit genes
Thirty-eight V. cholerae O1 strains, which include 32 from Zambia, one El Tor variant strain and three clinical hybrid strains (Matlab types I, II, III) from Bangladesh [Reference Nair13], one hybrid strain from Mozambique and reference strains of classical and El Tor biotypes were analysed by PFGE using the NotI restriction enzyme. The NotI restriction enzyme restricted the chromosomal genome to 15–27 fragments (Fig. 1) and the fragments ranged from 22·5 to 398 kb. Two major pulsotypes among the Zambian isolates, namely A and B, were observed.

Fig. 1. Dendrogram constructed from the PFGE profiles generated by NotI-digested genomic DNA of representative strains of V. cholerae O1 isolated in Zambia, hybrid strains from Bangladesh and Mozambique, and classical, El Tor reference strains.
Dendogram analysis showed that two major clusters, A and B, were produced by all strains isolated in Zambia. The El Tor variant strains from Bangladesh clustered together with strains isolated in Zambia in 2003 and 2004 (cluster B) with a similarity level of >0·98% indicating the same clonal origin (Fig. 1). The strains isolated in Zambia in 1996/1997 also produced a second cluster A with a similarity level of >0·98·5% but closely related to El Tor reference strain N16961, a Bangladeshi seventh-pandemic El Tor strain isolated in 1971 (Fig. 1). Second, hybrid El Tor strains from Matlab, Bangladesh (MJ-1236) [Reference Nair13] and Mozambique (B-33) carrying the classical CTX prophage [Reference Ansaruzzaman8] clustered closely (similarity level >0·90%). Finally, the Matlab variant strains MG-116226 (Matlab type II) and MG-116955 (Matlab type III), which are phenotypically different from conventional El Tor biotype of reference strain N16961, clustered together but quite differently from classical and El Tor reference strain O395 (similarity level <0·75%) and N16961 (similarity level <0·85%). The PFGE dendogram analysis showed that the strains isolated in Zambia between 2003 and 2004 were a different clonal cluster than that of strains isolated between 1996 and 1997. This result indicates that the new strains that emerged in 2003–2004 with a deletion of two ORFs, VC0493 and VC0498 of the VSP-II gene cluster was a recent occasion that may be clonally originated from the progenitor strain V. cholerae O1 El Tor strain. It is important to perform whole genome sequencing to understand the evolutionary changes of these newly emerging strains that appeared globally. Therefore, discovering how all these changes occurred in the Zambian V. cholerae O1 strains isolated between 1996 and 2004, and fitted into environmental niches needs to be elucidated.
Given that there are differences between the classical and El Tor biotypes, the selection of this El Tor strain seems to indicate an evolutionary optimization of the El Tor biotype and could represent a new, more efficient emerging form of the El Tor biotype of V. cholerae O1. As this El Tor variant biotype strain cannot be differentiated from other El Tor strains by currently used bacteriological methods, a revision of methods is needed to track the spread of such strains. The implication of heterogeneity in the B subunit of CTs of altered El Tor strains also needs to be assessed from a vaccine and diagnostic perspective. Perhaps the new type of V. cholerae strains have arisen from V. cholerae O1 strains having typical seventh-pandemic El Tor backgrounds with the replacement of their ctxB gene driven by the selective pressure to survive and adapt better in the host's intestine and other environmental niches. These new El Tor variant strains of V. cholerae O1, not only outfitted the conventional V. cholerae El Tor strains, but were also stable in nature. Our molecular characterization showed the emergence of a new variant strain of V. cholerae O1 in Zambia during 2003–2004 and how this strains was related to other variant strains isolated in Bangladesh. A continually changing pattern of V. cholerae O1 has been observed during the last decade in epidemic-prone areas especially in Asia and Africa.
There exists a definite need to identify and track the mode of spread of these new varieties of V. cholerae O1 strains in the population by vigilant surveillance systems, as these strains possess great potential for a new pandemic.
ACKNOWLEDGEMENTS
The work was supported by the Program of Founding Research Center for Emerging and Reemerging Infectious Diseases, Ministry of Education, Culture, Sports, Science and Technology of Japan and International Centre for Diarrhoeal Disease Research, Bangladesh (ICDDR,B). ICDDR,B acknowledges the following donors which provided unrestricted support to the Centre's research efforts: Australian Agency for International Development (AusAID), Government of the People's Republic of Bangladesh, Canadian International Development Agency (CIDA), Swedish International Development Cooperation Agency (Sida), Swiss Agency for Development and Cooperation (SDC) and the Department for International Development, UK (DFID).
DECLARATION OF INTEREST
None.






