The increasing prevalence of obesity is a major health problem as obesity is associated with chronic diseases, which include type II diabetes and CVDReference Silventoinen, Sans, Tolonen, Monterde, Kuulasmaa, Kesteloot and Tuomilehto1. An excessive adipose tissue mass, the main feature of obesity, is the result of an energy intake that exceeds energy expenditure for longer periods. Yet the molecular mechanisms contributing to the development of obesity remain ill-defined. Fat mass generation depends on an increased volume of adipocytes (hypertrophy), the recruitment of new adipocytes (hyperplasia), decreased apoptosis of adipocytes or a combination of these processesReference Prins and O'Rahilly2. Obesity accompanied by metabolic disturbances (including insulin resistance, increased production of NEFA, leptin and TNFα) is typically associated with hypertrophy of the fat cellsReference Farnier, Krief, Blache, Diot-Dupuy, Mory, Ferre and Bazin3.
PPARγ, a member of the nuclear hormone receptor superfamily of transcription factors, is required for adipogenesis, which involves the differentiation of preadipocytes into fat-storing mature adipocytesReference Rosen, Sarraf, Troy, Bradwin, Moore, Milstone, Spiegelman and Mortensen4. PPARγ regulates the expression of numerous genes involved in fat storage, including adipocyte fatty acid-binding protein (aP2), lipoprotein lipase and CD36. In addition, PPARγ is involved in the regulation of insulin sensitivityReference Rosen, Sarraf, Troy, Bradwin, Moore, Milstone, Spiegelman and Mortensen4, Reference Evans, Barish and Wang5. The importance of PPARγ activity in the regulation of fat mass is emphasized by PPARγ mutations. Dominant-positive PPARγ mutations are associated with severe obesityReference Ristow, Muller-Wieland, Pfeiffer, Krone and Kahn6, while dominant-negative PPARγ mutations are associated with partial lipodystrophyReference Savage, Tan and Acerini7. Activation of DNA transcription by PPARγ requires the binding of a ligand, as well as the presence of several transcriptional regulators and cofactorsReference Miard and Fajas8. Natural ligands for PPARγ include PUFA and eicosanoidsReference Kliewer, Sundseth and Jones9; synthetic ligands include rosiglitazone (RSG), which is widely and effectively used in the treatment of type II diabetes mellitus for its insulin-sensitizing effectsReference Lehmann, Moore, Smith-Oliver, Wilkison, Willson and Kliewer10. Treatment with RSG and other thiazolidinediones is accompanied by weight gain in rodentsReference Chaput, Saladin, Silvestre and Edgar11, Reference Larsen, Jensen, Sorensen, Larsen, Vrang, Wulff and Wassermann12 and obese, diabetic patientsReference Asnani, Richard, Desouza and Fonseca13. In rats, this is due to an increased food intake and an increased feed efficiencyReference Larsen, Jensen, Sorensen, Larsen, Vrang, Wulff and Wassermann12. However, the molecular and metabolic basis for the weight gain in obese diabetic patients is not completely clear. We recently overfed healthy, non-obese females and our results suggested that the ability to increase PPARγ activity may be involved in the susceptibility to gain weightReference Joosen, Bakker, Zorenc, Kersten, Schrauwen and Westerterp14. As PPARγ activity depends on ligand binding, the availability of ligand might be involved.
RSG appears to have its main effects on the adipose tissue, as RSG does only improve glucose and insulin levels in lipoatrophic, diabetic mice after fat transplantationReference Chao, Marcus-Samuels, Mason, Moitra, Vinson, Arioglu, Gavrilova and Reitman15. Specifically, RSG seems to affect the subcutaneous adipose tissue as only human subcutaneous, but not visceral, preadipocytes could be differentiated in vitro with RSGReference Hutley, Newell, Joyner, Suchting, Herington, Cameron and Prins16 and RSG increased subcutaneous, but not visceral, white adipose tissue in ratsReference Berthiaume, Sell, Lalonde, Gelinas, Tchernof, Richard and Deshaies17. In vivo, RSG has been shown to increase the expression of PPARγ response genes in type II diabetic patientsReference Tiikkainen, Hakkinen, Korsheninnikova, Nyman, Makimattila and Yki-Jarvinen18. We hypothesized that if PPARγ ligand levels are rate limiting in fat mass generation during overeating, RSG supplementation will increase subcutaneous fat storage. To address this question, healthy, non-obese males were exposed to an obesity-promoting environment while receiving either a placebo or RSG and the amount of TAG stored per fat cell, adipose gene expression, hormones involved in fat storage (insulin, leptin) and substrate availability (glucose, TAG) were determined.
Subjects and methods
Subjects
The present study was part of a study investigating the effects of the PPARγ ligand RSG on energy balance regulation, which is described previouslyReference Joosen, Bakker, Gering and Westerterp19. Subjects had to be male, between the age of 18 and 40 years, Caucasian, healthy and have a BMI between 20 and 32 kg/m2. They had to be unrestrained eaters, i.e. no desire to restrain intake, as indicated by scores ≤ 9 on factor 1 of the Three-Factor Eating Questionnaire, which reflects the extent to which individuals try to cognitively control their food intakeReference Stunkard and Messick20. Subjects completed a medical questionnaire before entering the study; only subjects in good health were included. Subjects who followed a dietary regimen with the aim to lose or gain weight within 1 year prior to the study were excluded. The study design required that the subjects were not fully informed about the adipogenic effect of the PPARγ agonist RSG as this could influence self-selected food and energy intake and activity-induced energy expenditure. Therefore, subjects were informed that they would receive either a substance that influences fat metabolism or a non-active substance (placebo) in a double-blind manner (i.e. neither the subject nor the investigator knew what the subject received during the experiment). The study was approved by the Ethics Committee of Maastricht University. All subjects received verbal and written information and signed a written consent form. Twenty men between the age of 20 and 29 years participated in the study. Characteristics of the subjects are shown in Table 1.
Table 1 Baseline characteristics of the subjects*
(Mean values with their standard errors)
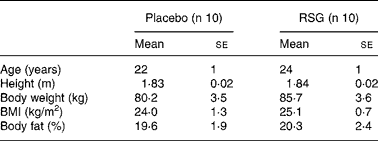
* For details of subjects and procedures, see Subjects and methods.
RSG, rosiglitazone.
Experimental design
Subjects were studied during a stay in the respiration chamber for 7 consecutive days for energy expenditure measurements calculated from VO2, CO2 production and urinary N excretion according to the formula of Brouwer Reference Brouwer21. The respiration chamber measures 14 m3 and is furnished with a bed, chair, table, TV, radio, telephone, computer, washbowl and toilet facilitiesReference Schoffelen, Westerterp, Saris and Ten Hoor22.
Following a double-blind, placebo-controlled design, subjects were randomly assigned to receive either 8 mg/d RSG (Avandia; GlaxoSmithKline BV, The Netherlands) or placebo orally. Drugs were dosed twice daily for the total stay of 7 d in the respiration chamber. Subjects were asked to eat ad libitum from an excess of food supplied at each mealtime and as snacks to induce a positive energy balance. No exercise protocol was imposed, but sleeping during daytime and strenuous physical activity were not allowed. Subjects were woken up between 08.00–08.30 hours, they were free to choose their bedtimes in the evening. Body composition was determined by underwater weighing and deuterium dilution (day 1) or by deuterium dilution alone (day 8). On the same days, blood samples and fat biopsies were taken. Fat storage was calculated as the difference between fat intake and fat oxidation.
Dietary intake
Meals consisting of typically Dutch food items were provided three times per d, breakfast between 08.30–09.00 hours, lunch between 12.30–13.00 hours and dinner between 18.00–18.30 hours. Breakfast and lunch consisted of bread, savoury and sweet condiments, fruit, yoghurt, milk, fruit juice, instant coffee (decaffeinated) and tea. For dinner, subjects could choose between ready prepared potato-, pasta- or rice-based meals with only vegetables or with vegetables plus meat or fish. They were allowed two alcoholic consumptions, white or red wine or beer, per d. A wide variety of snacks was continuously available in the chamber in packages that were replaced every morning. Snacks consisted of savoury items (crisps, nuts), sweet items (chocolate bars, sweets), various biscuits, fruit (apple, orange, banana), instant soup, fruit juice, instant coffee (decaffeinated) and tea. Meals and snacks were supplied in excess to be eaten ad libitum; extra food items were readily available on request. All foods and drinks entering and leaving the respiration chamber were weighed to the nearest gram. Energy content and macronutrient composition of the diets were calculated using the Dutch food composition table23.
Procedures
Anthropometry and body composition
Measurements were carried out in the morning after voiding and before breakfast. Body weight and height were measured to the nearest 0·01 kg and 0·1 cm respectively. BMI (kg/m2) was calculated as body weight (kg) divided by height (m) squared. Body density was determined by underwater weighing with simultaneous measurement of residual lung volume with the He dilution technique. Total body water was determined with deuterium dilution following the Maastricht protocolReference Westerterp, Wouters and Marken Lichtenbelt24. Body composition was calculated from body density and total body water using the three-compartment model of SiriReference Siri25.
Blood parameters
After an overnight fast, blood samples were obtained and mixed with citrate to prevent clotting. Plasma was obtained by centrifugation (4°C, 3000 rpm, 10 min), frozen in liquid N and stored at − 80°C until analysis of concentrations of glucose (hexokinase method, Glucose HK 125 kit; ABX diagnostics, Montpellier, France), insulin (ELISA, Mercodia, Uppsala, Sweden), leptin (double-antibody RIA, human leptin specific RIA kit, Linco Research Inc., St Charles, USA), TAG (Triglycerides liquicolor kit; Instruchemie, Delfzijl, The Netherlands), NEFA (Wako NEFA C-kit, Wako Chemicals, Neuss, Germany) and HDL-cholesterol (HDL cholesterol liquicolor kit; Instruchemie).
Adipose tissue and RNA isolation
Abdominal subcutaneous fat biopsies (approximately 500 mg) were obtained by needle liposuction under local anaesthesia (lidocain 2 %, AstraZeneca BV, Nederland) after an overnight fast. The tissue was immediately washed in cold PBS and cut into pieces with a scalpel. Then, tissue for mRNA analysis was immediately homogenized in 1 ml Trizol (Invitrogen Life Technologies, Carlsbad, CA, USA) in a mini-beadbeater (Biospec Products, Bartlesville, OK, USA), frozen in liquid N and stored at − 80°C until RNA extraction. Adipocytes were isolated from the remaining fresh tissue as described beforeReference Bakker, Van Dielen, Greve, Adam and Buurman26. Briefly, the pieces of fat tissue were digested in 5 ml Dulbecco's modified Eagle's medium (DMEM/F12; Invitrogen Life Technologies) containing 4 % bovine serum albumin (Sigma-Aldrich Co, St Louis, MO, USA) and 2 mg/ml collagenase II (Sigma) in an incubator (37°C, 5 % CO2) on a shaking platform (200 rpm) for 120 min. The digest was transferred to a 5 ml syringe and gently pressed over a 500 μm sterile disposable filter (Schleicher & Schuell MicroScience, Dassel, Germany). The syringe was rinsed with 5 ml DMEM/F12 containing 4 % bovine serum albumin, which was added to the digest. After centrifugation (1000 rpm, 1 min), the layer of adipocytes floating on top was removed, frozen in liquid N and stored at − 80°C until analysis of cell size by means of TAG and DNA content.
Total RNA was isolated using the method of Chomczynski and SacchiReference Chomczynski and Sacchi27 and 10 μg glycogen (Roche Diagnostics GmbH, Mannheim, Germany) was added to each sample. Extracted RNA was quantified and assessed for purity using the NanoDrop (NanoDrop Technologies, Wilmington, DE, USA) and gel electrophoresis (Agilent Technologies, The Netherlands).
Quantification of mRNA expression by real-time RT-PCR
cDNA synthesis and PCR reactions were performed as described previouslyReference Joosen, Bakker, Zorenc, Kersten, Schrauwen and Westerterp14. The primer sequences were as follows: PPARγ forward 5′-TCCATGCTGTTATGGGTGAA-3′, reverse 5′-TCAAAGGAGTGGGAGTGGTC-3′; aP2 forward 5′-GCATTCCACCACCAGTTTATC-3′, reverse 5′-CAGGAAAGTCAAGAGCACCAT-3′; fasting-induced adipose factor (FIAF) forward 5′- CACAGCCTGCAGACACAACTC-3′, reverse 5′-GGAGGCCAAACTGGCTTTGC-3′; adipsin forward 5′- TAGCGCGACTCCATCTCTACAA-3′, reverse 5′-GCCTCCTGAGTAGCTGGAACT-3′; adiponectin forward 5′-TATCCCCAACATGCCCATTCG-3′, reverse 5′-TGGTAGGCAAAGTAGTACAGCC-3′; 36B4 forward 5′- CGGGAAGGCTGTGGTGCTG-3′, reverse 5′-GTGAACACAAAGCCCACATTCC-3′; β-actin forward 5′-AGAAAATCT-GGCACCACACC-3′, reverse 5′-AGAGGCGTACAGGGATAGCA-3′. For target and housekeeping genes, standard curves were created from a specific PCR productReference Joosen, Bakker, Zorenc, Kersten, Schrauwen and Westerterp14. To account for differences in RNA loading, target mRNA was expressed relative to β-actin and relative to 36B4 mRNA. As both housekeeping genes gave the same normalized quantities, target mRNA levels are presented after normalization to β-actin.
For determination of TAG and DNA in adipocytes, about 40 mg adipocytes was incubated in a volume of 160 μl containing 0·1 m-NaCl, 0·02 m-Tris pH 7·4, 0·01 m-EDTA pH 7·5, 100 μg/ml proteinase K and 0·05 % Triton X-100 for 60 min at 65°C, followed by 10 min at 95°C. Of this solution, 10 μl was 100 × diluted before determination of TAG (GPO-trinder 337; Sigma). For DNA determination, the remaining solution was mixed with SDS, RNase A and proteinase K to make final concentrations of 0·5 %, 20 μg/ml and 100 μg/ml respectively. After an overnight incubation at 37°C, DNA was precipitated using ethanol precipitation and a high-salt solution (Kac), rinsed with 70 % ethanol and resuspended in 20 μl H2O. DNA was quantified and assessed for purity using the NanoDrop (NanoDrop Technologies). The mean adipocyte cell size was calculated as μg TAG/cell (mean DNA content per cell is 6 pg).
Statistical analysis
Results are presented as means with their standard errors or as median (inter-quartile range). Measurements on the morning of day 1 before entering the respiration chamber are referred to as baseline; measurements on the morning of day 8 after the 7-d respiration chamber stay are referred to as overeating. Effects of overeating within groups were analysed with Student's paired t test (two-sided) or Wilcoxon's paired signed rank test (two-sided) where appropriate. Effects of RSG treatment (between-group effects) were analysed with the Mann–Whitney U -test (two-sided). Significance of correlations was tested with the Spearman rank correlation (r S). P < 0·05 was considered statistically significant. SPSS 11 for Macintosh (SPSS Inc., Chicago, IL, USA) was used for the analysis.
Results
Obesity-promoting environment
For both groups, energy intake (placebo 15·9 (se 0·9) MJ/d v. RSG 18·9 (se 1·2) MJ/d, P = 0·06) was higher than energy expenditure (placebo 11·3 (se 0·3) MJ/d v. RSG 12·5 (se 0·5) MJ/d, P = 0·09). By the end of day 7, the cumulative energy balances were 32·2 (se 5·1) MJ and 44·7 (se 6·9) MJ for the placebo and the RSG groups respectively, which was not statistically significant different due to the high inter-individual variation in both groups (P = 0·16). Body weight increased by 1·39 (se 0·37) kg (P < 0·01) in the placebo group and by 2·54 (se 0·53) kg (P = 0·001) in the RSG group. Changes in body weight did not differ significantly between treatments (P = 0·09). However, body weight change on RSG was accompanied with an increase in total body water of 1·6 (se 0·6) litres (P < 0·05), whereas total body water did not significantly change in the placebo group (–0·2 (se 0·7) litres, P = 0·79).
Rosiglitazone causes elevated TAG levels
In the placebo group, plasma glucose and NEFA were decreased and TAG was increased after overeating, other parameters did not change significantly (Table 2). In the RSG group, plasma insulin and TAG were increased and NEFA decreased after overeating; plasma glucose, leptin and HDL-cholesterol did not change significantly. RSG affected changes in plasma glucose (P = 0·02) and TAG (P = 0·03), but not in plasma NEFA (P = 0·94), insulin (P = 0·07), leptin (P = 0·15) and HDL-cholesterol (P = 0·97). The difference in baseline insulin levels between groups was not statistically significant (P = 0·11).
Table 2 Fasting plasma parameters at baseline and after self-induced overeating for the placebo and rosiglitazone (RSG)-treated groups‡
(Mean values with their standard errors)

Overeating significantly different from baseline (within-group) using Wilcoxon's paired signed rank test: *P < 0·05, **P < 0·01, ††P = 0·01.
Change in time significantly different between the placebo and RSG-treated groups using the Mann–Whitney U -test: †P < 0·05.
‡ For details of subjects and procedures, see Subjects and methods.
Rosiglitazone has no effect on PPARγ transcriptional activity in subcutaneous adipose tissue
The measured mRNA levels represent PPARγ response genes involved in adipogenesis, lipogenesis and lipolysis, i.e. PPARγ, aP2, adipsin, adiponectin and FIAF (Table 3). In the RSG group, FIAF mRNA levels were significantly decreased after overeating (median fold change 0·30, P = 0·01), with a trend towards a decrease in the placebo group. However, transcript levels were not statistically significant affected by RSG treatment.
Table 3 Median (inter-quartile range; IQR) fold changes in mRNA expression in subcutaneous abdominal adipose tissue after self-induced overeating for the placebo and rosiglitazone (RSG)-treated groups‡
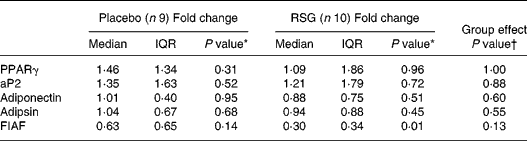
* Using Wilcoxon's paired signed rank test within groups.
† Using the Mann–Whitney U -test between groups.
‡ For details of subjects and procedures, see Subjects and methods.
aP2, adipocyte fatty acid-binding protein; FIAF, fasting-induced adipose factor.
In both groups, there were strong correlations between PPARγ and aP2 mRNA at baseline, after overeating and between changes in expression (r S 0·83–0·97, P < 0·01), indicating that PPARγ mRNA levels are related to PPARγ activity. There were no significant correlations between changes in PPARγ or aP2 mRNA levels and fat storage. Changes in adiponectin mRNA levels were positively correlated with changes in PPARγ or aP2 mRNA levels in both groups (r S 0·64–0·93, P < 0·05).
Rosiglitazone does not affect fat storage strategy
The median fat cell size decreased with 50 % in both the placebo and the RSG groups, but this change was not statistically significant (P = 0·11 and 0·08 respectively; Table 4 and Fig. 1). There was no statistically significant difference in changes in fat cell size between treatments (P = 0·40); for the entire group, fat cell size was decreased after overeating (P = 0·02). These changes were accompanied by a, not statistically significant, increase in DNA content (P = 0·77 placebo; P = 0·14 RSG). There were no differences in gene expression or plasma parameters between subjects who showed fat cell hypertrophy or hyperplasia after overeating for both groups.
Table 4 TAG and DNA contents of subcutaneous adipocytes at baseline and after self-induced overeating for the placebo and rosiglitazone (RSG)-treated groups†

* Calculated from the mean DNA content per cell; 6 pg.
† For details of subjects and procedures, see Subjects and methods.
IQR, inter-quartile range.
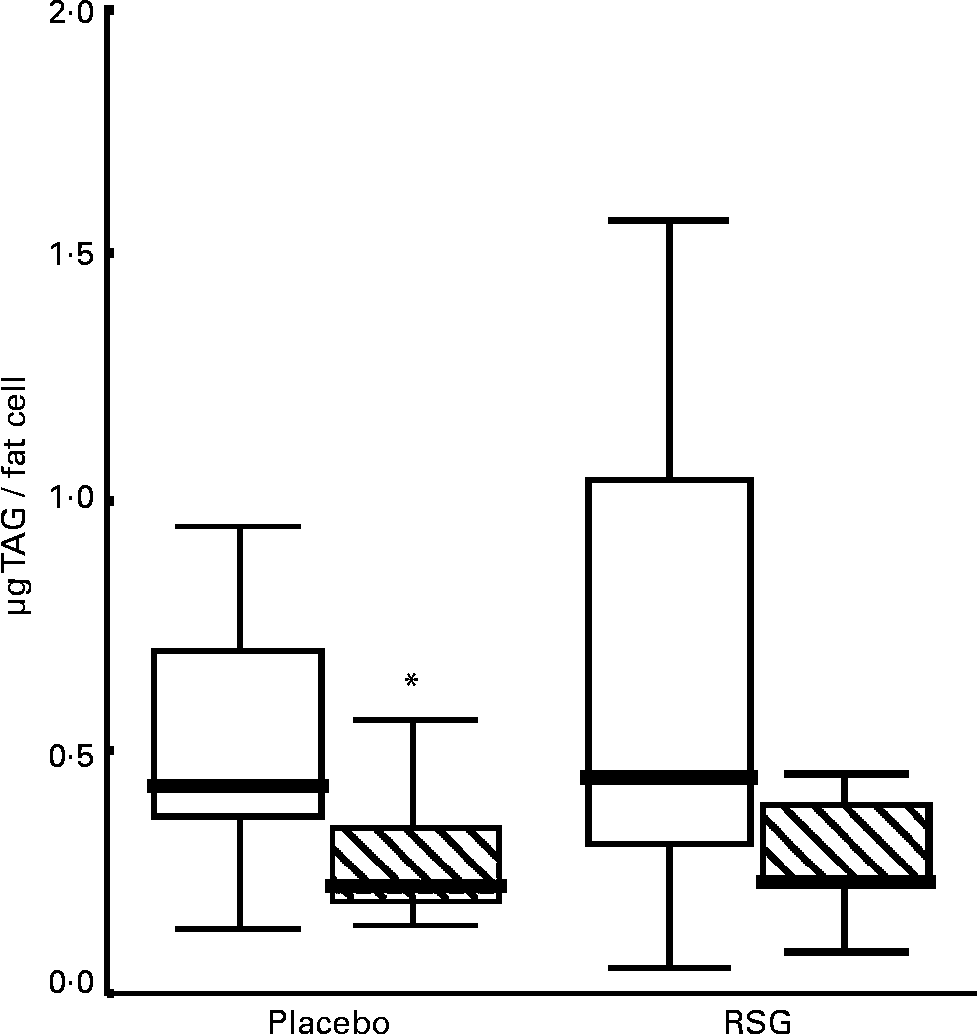
Fig. 1 Boxplot graph of fat cell size in subcutaneous abdominal adipose tissue at baseline (□) and after self-induced overeating (▧) (n 9 for each treatment due to small fat biopsies in two subjects). Outlier marked with an asterisk. RSG, rosiglitazone. For details of subjects and procedures, see Subjects and methods.
Discussion
PPARγ is the key regulator of adipose gene expression, whose activity depends on transcriptional regulators and cofactors as well as on ligand availabilityReference Miard and Fajas8. To study whether the availability of PPARγ ligand is rate limiting in fat mass generation, we studied the effects of RSG supplementation on several PPARγ-dependent processes. The results indicate that ligand availability is of minor importance in subcutaneous fat mass generation.
In both the placebo and RSG groups, PPARγ and aP2 mRNA levels were correlated, which indicates activity of PPARγ as aP2 is primarily a PPARγ response geneReference Ross, Graves, Greenstein, Platt, Shyu, Mellovitz and Spiegelman28, Reference Sen, Lea-Currie, Sujkowska, Franklin, Wilkison, Halvorsen and Gimble29. However, aP2 gene expression was not different between treatments, suggesting that PPARγ ligand availability is not rate limiting for PPARγ activity in subcutaneous adipose tissue in healthy male subjects. Similar results were shown in type II diabetic patients treated with pioglitazone for 24 weeksReference Bogacka, Xie, Bray and Smith30. Pioglitazone treatment did not change PPARγ mRNA levels and protein content, but baseline levels were strongly correlated with the expression genes involved in fatty acid metabolism, synthesis and storage (acetyl-CoA synthetase, lipoprotein lipase and fatty acid synthase)Reference Bogacka, Xie, Bray and Smith30. Unfortunately, because of the small fat biopsies, we were not able to determine protein levels.
Adiponectin and adipsin are respectively early and late markers of adipocyte differentiationReference Gustafson, Jack, Cushman and Smith31, Reference Xia and Cianflone32. While plasma adipsin concentrations are increased in obesity and correlate with plasma insulin and TAG, adipsin mRNA in subcutaneous fat tissue decreases with increasing BMIReference Xia and Cianflone32. In contrast, plasma adiponectin and mRNA expression are decreased in obesity and in insulin-resistant patients, independent of obesityReference Gustafson, Jack, Cushman and Smith31 and can be increased by thiazolidinedione treatmentReference Maeda, Takahashi and Funahashi33. These observations suggest an impaired fat cell function due to a diabetic state, which is typically associated with fat cell hypertrophyReference Farnier, Krief, Blache, Diot-Dupuy, Mory, Ferre and Bazin3. Indeed, fat cell size was shown to be negatively correlated with adiponectin mRNAReference Yang, Jansson, Nagaev, Jack, Carvalho, Sunnerhagen, Cam, Cushman and Smith34. However, we found no effect of RSG or overeating on adiponectin and adipsin mRNA expression, suggesting that RSG has no short-term, or at least an insignificant short-term, effect on adipose gene expression in healthy, non-obese males.
Interestingly, RSG had a significant effect on TAG metabolism (i.e. tissue clearance and/or endogenous synthesis) above the direct postprandial changes in substrate metabolism, as the increase in fasting plasma TAG concentration after overeating was significantly higher in the RSG group. Although carbohydrate intake was higher in the RSG groupReference Joosen, Bakker, Gering and Westerterp19, we believe this is not due to differences in nutrient-related (i.e. fructose) TAG production as fruit intake was not significantly different (data not shown). However, we showed previously that whole-body fat oxidation was more suppressed in the RSG groupReference Joosen, Bakker, Gering and Westerterp19 indicating that the increase in TAG levels resulted from lowered NEFA oxidation in the muscles while physical activity levels were not significantly different between groups (data not shown). These results are in strong contrast to the effects of RSG on the adipose tissue of diabetic patientsReference Boden, Homko, Mozzoli, Showe, Nichols and Cheung35. A factor directly related to plasma TAG levels and positively regulated by PPARγ is FIAF, which is primarily produced by the adipose tissue and owes its name to its strong up regulation in adipose tissue and liver during fastingReference Kersten, Mandard, Tan, Escher, Metzger, Chambon, Gonzalez, Desvergne and Wahli36. Prolonged high-fat feeding in mice lowers plasma FIAF levels, but not adipose tissue mRNA, and increases plasma TAG levels probably by inhibiting lipoprotein lipase activityReference Kersten, Mandard, Tan, Escher, Metzger, Chambon, Gonzalez, Desvergne and Wahli36, Reference Mandard, Zandbergen and Tan37. We did not find a statistically significant difference in FIAF mRNA between groups, but as adipose FIAF expression decreased during overeating in the RSG group, with a tendency towards a decrease in the placebo group, FIAF may be involved in the observed effects of RSG on TAG metabolism.
In addition to influencing TAG metabolism, RSG treatment was accompanied by pronounced fluid retention. Recent studies in mice show that PPARγ has a direct role in renal Na re-absorption, providing a mechanism for RSG-induced fluid retentionReference Guan, Hao and Cha38, Reference Zhang, Zhang, Kohan, Nelson, Gonzalez and Yang39. In addition, RSG increased weight and vascular permeability selectively in adipose tissue, but not in skeletal muscle, in lean, fatty and diabetic fatty Zucker rats, indicating that insulin sensitivity is not essential for RSG to induce fluid retentionReference Sotiropoulos, Clermont and Yasuda40.
In mice, high PPARγ activity is associated with an increase in fat cell size on a high-fat diet, while low PPARγ activity is associated with an increase in fat cell numberReference Kubota, Terauchi and Miki41. In the present study, fat cell size was decreased after overeating, suggesting that fat was stored in newly recruited fat cells. RSG did not influence this storage strategy.
We can speculate that the fat mass itself might be involved in ligand availability, ligand binding and cofactor recruitment, which might explain why RSG has different effects in non-obese, healthy subjects compared with obese, diabetic patients. Furthermore, despite the absence of increased transcriptional activity of PPARγ in the subcutaneous adipose tissue, the primary target tissue of RSG, RSG induced pronounced changes in water balance and fat oxidation and tended to increase food intakeReference Joosen, Bakker, Gering and Westerterp19. Although effects on transcriptional activity in other organs cannot be ruled out, the adipose tissue is widely recognized as the main site of action of PPARγ agonists. In the absence of differences in PPARγ activity between treatments, alternative pathways for RSG may have been involved. In rats, thiazolidinediones acutely activate AMP-activated protein kinase in adipose tissue, and also in skeletal muscle and liver. This effect is most likely independent of PPARγ-activated gene transcriptionReference LeBrasseur, Kelly, Tsao, Farmer, Saha, Ruderman and Tomas42. In turn, phosphorylation of PPARγ by AMP-activated protein kinase inhibits its transcriptional activityReference Diradourian, Girard and Pegorier43. However, we can only speculate, as this experiment was not designed to study non-transcriptional effects of RSG. The possibility of alternative pathways and, if any, their relative importance and changes in time need to be investigated in further research.
In conclusion, the present results suggest that in healthy, non-obese males the PPARγ ligand RSG influences TAG metabolism without increasing the transcriptional activity of PPARγ in the subcutaneous adipose tissue.
Acknowledgements
We thank Maarten Gering for help in data collection and Gabby Hul for obtaining the fat samples. None of the authors had financial or personal conflicts of interest.







