1. Introduction
Dilated cardiomyopathy (DCM) is a cardiac muscle disorder that leads to cardiovascular morbidity and mortality. It is associated with heart failure and sudden cardiac death. This disorder is known to develop at any age, in either sex and in people of any ethnic origin (Taylor et al., Reference Taylor, Carniel and Mestroni2006). The prevalence of DCM in the general population of India is not well defined (Ushasree et al., Reference Ushasree, Shivani, Venkateshwari, Jain, Narsimhan and Nallari2009).
Over the past few years, studies on the application of genetic and genomic techniques to understand the aetiology of DCM have increased. With a focus on molecular diagnostics and genetic intervention, it is aiding in the improvement of clinical practice and routine management of the patients. Of the many approaches in genetics, studies on DNA damage remain an important aspect to understand the complete molecular nature of disorders. Very few studies have reported DNA damage in cardiovascular patients using DNA adducts and cytogenetic damage in peripheral blood lymphocytes as endpoints (Botto et al., Reference Botto, Rizza, Colombo, Mazzone, Manfredi, Masetti, Clerico, Biagini and Andreassi2001; Murgia et al., Reference Murgia, Maggini, Barale and Rossi2007). However, no data exist on DNA damage in lymphocytes of DCM patients. The cytokinesis-block micronucleus cytome (CBMN Cyt) assay is an efficient technique to assess cellular and nuclear malfunctions and dysfunctions by measuring the frequencies of MN and other genomic aberrations in cultured human cells (Fenech, Reference Fenech1997).
In this study, the possible effects of DNA damage on the development and progression of the disease were also investigated by mutation analysis of the cardiac Troponin C type I (TNNC1) gene. TNNC1 was identified as a novel gene for DCM in the study conducted by Mogensen et al. (Reference Mogensen, Murphy, Shaw, Bahl, Redwood, Watkins, Burke, Elliott and McKenna2004). A few studies have reported the presence of mutations in the gene TNNC1 coding for cTnC in DCM patients. The cTnC mutations E59D and D75Y were detected in only one patient with DCM. This raised the question of whether this mutation co-segregated with the disease. However, from functional studies it was identified that these mutations have a synergistic effect on Ca2+ sensitivity and ATPase activation.
The aim of the present study was to assess the DNA damage by employing CBMN Cyt assay in cultured peripheral blood lymphocytes of DCM patients. An effort was also made to understand if there was any association between DNA damage and disease progression.
2. Materials and methods
(i) Populations studied
Informed written consent was obtained from 48 patients suffering from DCM and undergoing treatment at Heartline Medical and Research Centre, Vellore, India and 48 age- and sex-matched controls (n = 48). Blood samples were collected by venipuncture in heparinized and EDTA-coated vacutainers.
(ii) Ethical standards
Ethical clearance for this study was obtained from the institutional review board of VIT University. The authors assert that all procedures contributing to this work comply with the ethical standards of the relevant national and institutional committees on human experimentation and with the Helsinki Declaration of 1975, as revised in 2008.
(iii) Clinical study
The clinical evaluation was based on the guidelines prescribed by Dickstein et al. (Reference Dickstein, Cohen-Solal, Filippatos, McMurray, Ponikowski, Poole-Wilson, Strömberg, van Veldhuisen, Atar, Hoes, Keren, Mebazaa, Nieminen, Priori and Swedberg2008). Clinical evaluation consisted of recording a careful patient and family history, electrocardiogram (ECG), echocardiogram, chest X-ray and routine blood chemistry. Based on the clinical presentation, the exclusion criteria were decided. Patients with known history of rheumatic heart disease, joint pain, chronic hypertension, anaemia and those having abnormal values for creatinine, urea and albumin were not included in the study.
DCM was confirmed using an M-Mode, 2D echocardiogram with colour Doppler, by the presence of left ventricular dilatation and dysfunction with ejection fraction (EF) <50% and fractional shortening (FS) <30, no region of wall abnormalities in the echo and normal ECG. The left ventricle dimensions were obtained using the Teichholz method. The frequencies of the clinical characteristics such as EF, FS, mitral regurgitation and tricuspid regurgitation were recorded.
(iv) Detailed history and physical examination
Alcohol use, cocaine use, dietary pattern, past illnesses (including infections involving the heart, history of arrhythmias, etc.), medications taken, pregnancies, occupations of the patients and family history were recoded. The occupational characteristics of the patients and the controls were recorded, to account for exposures to toxins and mutagens, if any.
(v) Cytogenetic studies
Peripheral blood samples were processed immediately on sampling and cultured. Chromosome preparations were made according to the standard procedure (Hungerford, Reference Hungerford1965). Cultures were set up by mixing 0·5 ml of whole blood with 6 ml of Ham's F10 media (PAN Biotech GmBH), supplemented with 1·2 ml of fetal bovine serum (Hi Media) and 0·3 ml of phytohaemagglutinin (GIBCO).
(a) CBMN Cyt assay
For this assay, lymphocyte cultures were set up as above. Cells were blocked in cytokinesis at the 44th hour by the addition of Cytochalasin B (6 μg/ml final concentration, Sigma). Harvesting was carried out at the end of 72 h. The cells were treated hypotonically with 0·075 m KCl for 8 min, followed by fixation with methanol–acetic acid (3 : 1). Subsequently, slides were prepared and stained with Giemsa solution (4%). For each sample, 1000 binucleated cells were scored for abnormalities following the criteria specified by Fenech et al. (Reference Fenech, Chang, Kirsch-Volders, Holland, Bonassi and Zeiger2003).
(vi) Molecular studies
Blood samples collected in EDTA vacutainers were processed for DNA extraction. Total genomic DNA was extracted from 100 μl peripheral blood samples from patients using DNeasy blood & tissue kit (Qiagen, Germany). Primers were designed and the standard PCR reaction for a 20 μl reaction mixture was carried out. PCR products were analysed by agarose gel electrophoresis. PCR products were purified using MinElute PCR Purification Kit (Qiagen, Germany). Purified PCR products were quantified in synergy H4 multimode micro-plate reader using Take3 micro-volume plate. The purified exon 4 amplified PCR product was cycle sequenced using BigDye terminator (Life Technologies, USA), and the labelling reaction was carried out from both ends using the forward and reverse primers individually. The PCR products were purified by Ethanol/EDTA precipitation method. The purified product was denatured using 10 μl of Hi-Di Formamide and it was run on a AB3730 DNA analyser. The raw data files were analysed using Chromas lite Software (Technelysium, USA) to detect for any mutation in the patient sample compared with the normal sequence of TNNCI gene.
(vii) Statistical analysis
A comparison between the data obtained from the patient group (n = 48) and the control group (n = 48) was analysed using the t test. A P value of <0·001 with confidence limit of 95% was defined as statistically significant. Numerical data are presented as mean ± sd Microsoft Excel and WinStat were used for the statistical analysis.
3. Results
No significant difference was observed in the demographic profile and the lifestyle of the patient group and the control group. The individuals studied had varied occupation. The basic diet of both the patient group and control group was a typical South Indian diet.
(i) CBMN Cyt assay
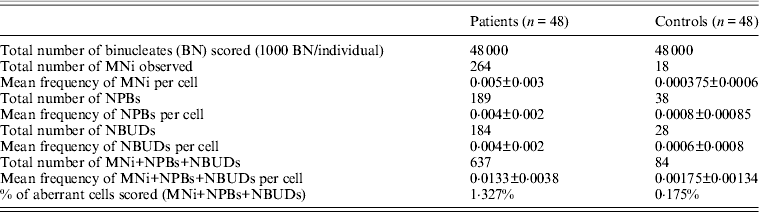
A significantly higher frequency of micronuclei (MNi) was observed in the patient group than that of the control group (P<0·001) (Table 1). The mean frequency of MNi per cell observed for the patient group was 0·005±0·003 and for the control group was 0·000375±0·0006 (P<0·001). A significant number of nuclear buds (NBUDS) were also observed in the patient group (0·004±0·002 per cell), as compared with the control group (0·0006±0·0008 per cell) (P<0·001). A higher frequency of nucleoplasmic bridges (NPBs) in the patients (0·004±0·002 per cell) was observed as compared with the controls (0·0008±0·00085 per cell) (P<0·001). No significant difference was observed in the mean frequencies of aberrations detected by the CBMN Cyt assay in the patient group due to age, gender, smoking, alcohol drinking, dietary patterns or their occupation (P>0·005). The frequency of anomalies grouped according to disease severity, established by EF measurement showed no differences. The frequency distributions of various anomalies detected by the CBMN Cyt assay, in the control group and patient group are presented in Fig. 1. All patients had at least three to five aberrations. Seventeen patients had 12–14 aberrations. In one patient, 26 aberrations were scored. Whereas in the control group, 12 individuals had three to five aberrations and 36 individuals had only one aberration. The ratio of binucleated cells to mononucleated cells was calculated as 1·04 for patients and 1·01 for controls. No significant difference was found in the ratio of binucleated cells to mononucleated cells in the patient and control groups. The percentage of aberrant cells containing MNi with NBUDS and NPBs in the patient group is 1·327%, whereas in the control group it is 0·175%.
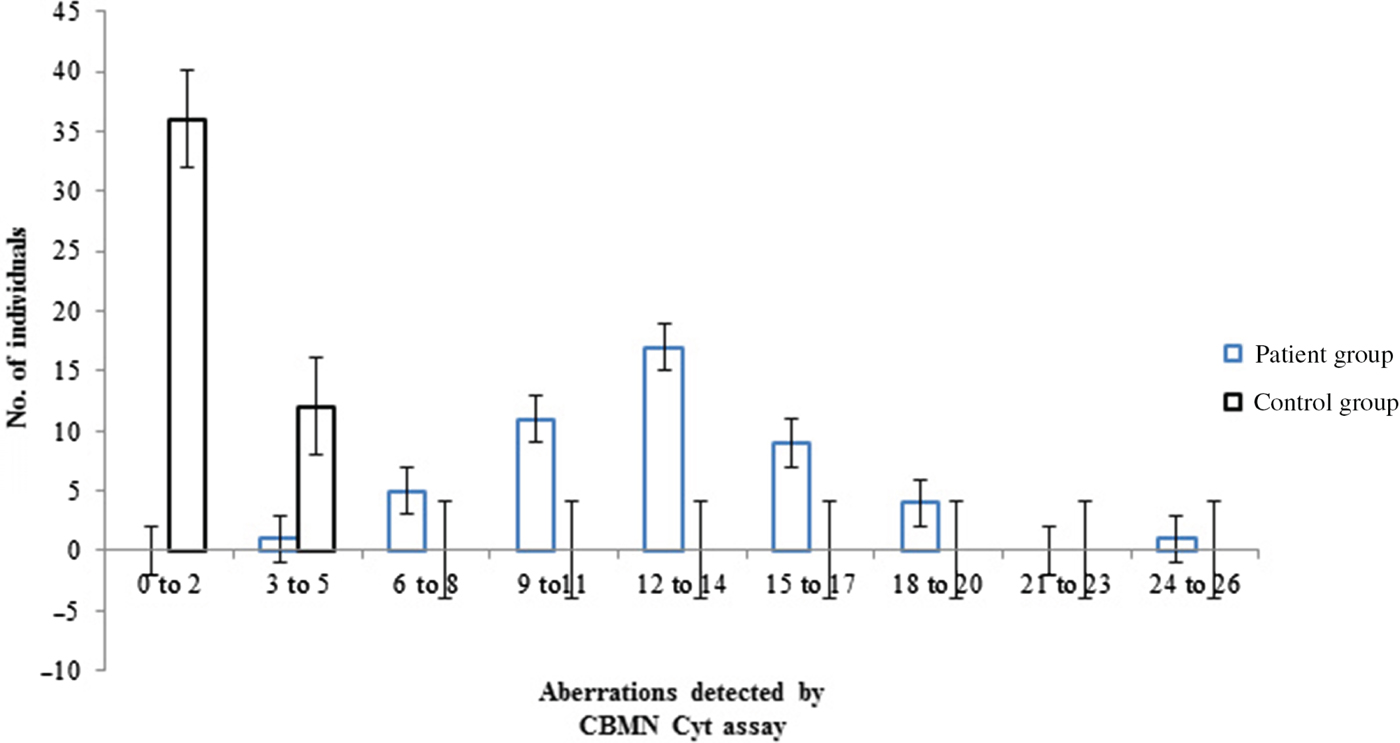
Fig. 1. Frequency distribution of aberrations detected by CBMN Cyt assay in patients.
Table 1. Results of the CBMN Cyt assay
(ii) Mutation analysis
The primers were designed approximately 100 base pairs up-stream and down-stream of exon 4 with the forward primer sequence of 5′-CTCCCCAGGCTCCAGAAG-3′ and reverse primer sequence 5′-GCATCCCTCTCCCCTATCAG-3′. Sequence data analysis revealed no variations in the gene sequence of the patients and the normal TNNC1 gene sequence.
4. Discussion
Two main causes of death in the global population are cardiovascular diseases and cancer. It is hypothesized that these chronic degenerative diseases share several risk factors, such as oxidative stress and DNA damage (Trosko & Chan, Reference Trosko and Chan1980). Although the role of DNA damage and mutation in carcinogenesis is well established, their involvement in vascular diseases is less understood.
In general, in any biological system for the detection and estimation of naturally occurring and induced genotoxic effects, either chromosomal aberration (CA) frequencies or MNi frequencies are used. In an earlier study, we reported high frequency of DNA damage as expressed in various types of CA such as chromosomal breaks, chromatid breaks and dicentric chromosomes (Sitaraman et al., Reference Sitaraman, Babu and Saraswathy2012). It was observed in that study that the mean value of all CAs was 0·092 ± 0·04. In the present study, through the application of CBMN Cyt assay, the NPBs between the binucleate cells (0·004 ± 0·002) and nuclear budding (0·004 ± 0·002) gave added informational parameters along with the frequency of MNi. It is noted that although the frequencies of CAs, as reported previously and the various anomalies detected by the CBMN Cyt assay are comparable, the frequency of CA per cell (0·092 ± 0·04) is higher than the frequency of MNi per cell (0·0133 ± 0·0038).
There was no significant difference between the ratio of binucleated cells to mononucleated cells in the control and patient groups, produced by the CBMN Cyt assay. This indicates that the cell cycle progression in the patient group is similar to that of the control group and is not affected by any extrinsic or intrinsic factor.
The other genomic aberrations as seen in this study may help to have a clearer picture of the complete DNA damage in the DCM syndrome. NBUDS are known to form due to the elimination of amplified DNA or sequence to distinct region of the nucleus, usually the nuclear periphery. NBUDS have also been shown to be formed when an NPB between two nuclei breaks and the remnants shrink back towards the nuclei (Fenech, Reference Fenech2007; Fenech et al., Reference Fenech, Kirsch-Volders, Natarajan, Surralles, Crott, Parry, Norppa, Eastmond, Tucker and Thomas2011).
No mutations were identified in the TNNC1 gene. Hence, it is not clear from this study whether mutations may arise as a consequence of DNA damage. Although a few studies have reported functional studies on mutations in cTnC and have shown the effect of cTnC mutations in muscle contractility (Mogensen et al., Reference Mogensen, Murphy, Shaw, Bahl, Redwood, Watkins, Burke, Elliott and McKenna2004; Hershberger et al., Reference Hershberger, Norton, Morales, Li, Siegfried and Gonzalez-Quintana2010; Pinto et al., Reference Pinto, Siegfried, Parvatiyar, Li, Norton, Jones, Liang, Potter and Hershberger2011), it is highly likely that cTnC being the Ca2+ sensor of cardiac muscle, any mutations in this protein might be lethal and therefore possibly be non-existent in the adult population. It is also possible that the mutations exist but have no overall effect on cardiac function.
It was also observed that no correlation could be established between the severity of the phenotype, as expressed as lower EF and DNA damage measured. Hence, the high frequency of aberrations observed in the patient group may be attributed to co-existing factors of the disease rather than the disease itself. It is hypothesized that psychological stress combined with the clinical presentation of the disease may have led to a higher frequency of DNA damage. Several studies have reported that stress can be responsible for induction of DNA damage (Fischman & Kelly, Reference Fischman and Kelly1999; Dimitroglou et al., Reference Dimitroglou, Zafiropolou, Nikolaki, Doudounakis, Tsilimigaki and Piperakis2003).
Although in this study significant DNA damage in the lymphocytes of the patients has been observed, it is important to note that given the small size of the cohort, the results may not be representative over a generalized patient population. Studies on larger cohorts may help in establishing the effect that DNA damage has on the functional unit of the muscle, which is seen to be affected in DCM.
The authors would like to sincerely thank the patients who participated in this study. We wish to acknowledge VIT University for providing the necessary facilities to carry out this study. SS is grateful for the Research Associateship provided by the VIT University during her study.
Financial Support
This research received no specific grant from any funding agency, commercial or not-for-profit sectors.
Statement of Interest
None.




