A recent meta-analysis of prospective cohort studies shows that dietary linoleic acid (LA) is inversely associated with CHD risk in a dose-response manner(Reference Farvid, Ding and Pan1). However, as an essential fatty acid, the health effects of circulating LA remain unclear. The objective of the study was to explore the association between plasma LA and markers of metabolic health, and to identify key determinants of plasma LA.
Plasma fatty acids in 820 fasting blood samples from the Irish National Adult Nutrition Survey(Reference Li, Brennan and McNulty2) were extracted, trans-esterified and determined using GC-MS(Reference Li, Brennan and McNulty2). 15 SNPs of the FADS1/2 gene were examined. Linkage disequilibrium and haplotypes were analysed using HaploView and R package Haplo.stat respectively. Haplotype effects were estimated using HapStat 3·0(Reference Lin, Zeng and Millikan3). Differences in demographics, dietary intakes, markers of metabolic health were compared using GLM adjusted for confounders across plasma LA quartiles using SPSS.
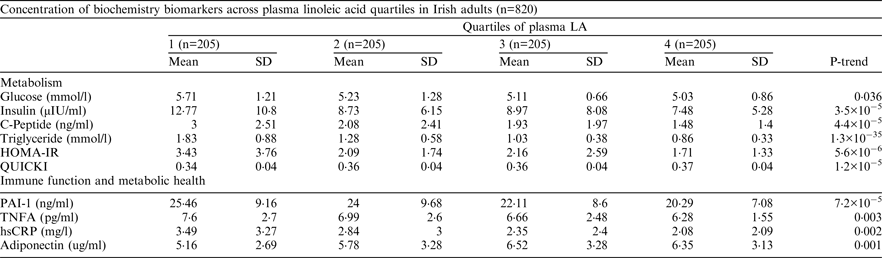
Comparisons of concentration of biochemical biomarkers and plasma fatty acids (log transformed for skewed valuable) across quartiles were assessed using General Linear Contrast Model adjusted for age, body fat, blood pressure, waist circumference, waist hip ratio, smoking status, working energy expenditure and BMI.
P-values were corrected by multiple number of traits. P-values that exceed 1·0 after correction for multiple testing have been marked down to 1.000.
Subjects in the highest quartile of plasma LA showed healthier metabolic profile in terms of blood lipids, insulin sensitivity and inflammation markers (PAI-1, hs-CRP, etc) (P < 0·001). The main determinants of plasma LA were waist circumference (8·1 %), 15-locus haplotype (3·9 % of variance), dietary PUFA intakes (3·7 % of variance) and body fat (3·7 % of variance) (P < 0·001). There was no haplotype × dietary PUFA interaction after adjustment for confounders including covariates, haplotype × covariate and dietary PUFA × covariate.
Adiposity, dietary PUFA intakes and haplotypes were the main determinants of plasma LA level. Higher plasma LA level was linked to a healthier metabolic profile and lower metabolic risk.
The project was funded by the Department of Agriculture, Fisheries and Food and the Health Research Board under the Food for Health Research Initiative 2007–2012. Mr Li is in receipt of a PhD student from the China Scholarship Council 2012–2016.


