Globally, norovirus is a common cause of acute gastroenteritis [Reference Heymann1]. In 2010, it caused an estimated 97.6 cases of acute gastroenteritis per 1000 population in Australia [Reference Gibney2]. Although the symptoms of norovirus are generally mild and self-limiting, its high infectivity often results in widespread illness and outbreaks [Reference Heymann1, Reference White3].
Every 2–3 years antigenic shift or recombination has led to a new strain of norovirus emerging that replaces the current circulating strain, with resulting low population immunity [Reference White3]. Since 2012, the predominant pandemic strain has been GII.Pe/GII.4 Sydney 2012 [Reference White3]; however, in recent years several new norovirus strains have appeared as contenders for the next predominant strain [Reference Lu4–Reference Ruis8]. This paper describes an outbreak in a regional area of NSW caused by the newly emerging GII.P16/GII.4 Sydney 2012 recombinant virus.
On 11 August 2017, the Hunter New England public health unit in NSW was notified of several cases of gastroenteritis amongst attendees of a health department function held 3 days earlier. An event-specific online questionnaire investigating food exposures and symptom profile was developed and sent to 227 of the 240 function attendees whose email addresses were available. A total of 180 attendees (79%) completed the online survey with 32 classified as cases based on an event-specific case definition. Within 3 h of sending out the questionnaire, 92 attendees had responded, allowing for rapid epidemiological analysis and assessment of the situation. Two cases were laboratory confirmed with norovirus genotype GII.P16/GII.4 Sydney 2012, whilst the remaining 30 cases meeting the probable case definition (a person who experienced symptoms of diarrhoea ⩾3 or more loose stools) or vomiting after attendance at the event on 8 August 2017, with the onset of diarrhoea or vomiting occurring after the minimum norovirus incubation period of 10 h. An epidemic curve of the outbreak (Fig. 1) showed a typical point source pattern with a median incubation period of 33 h (range 12–61 h).
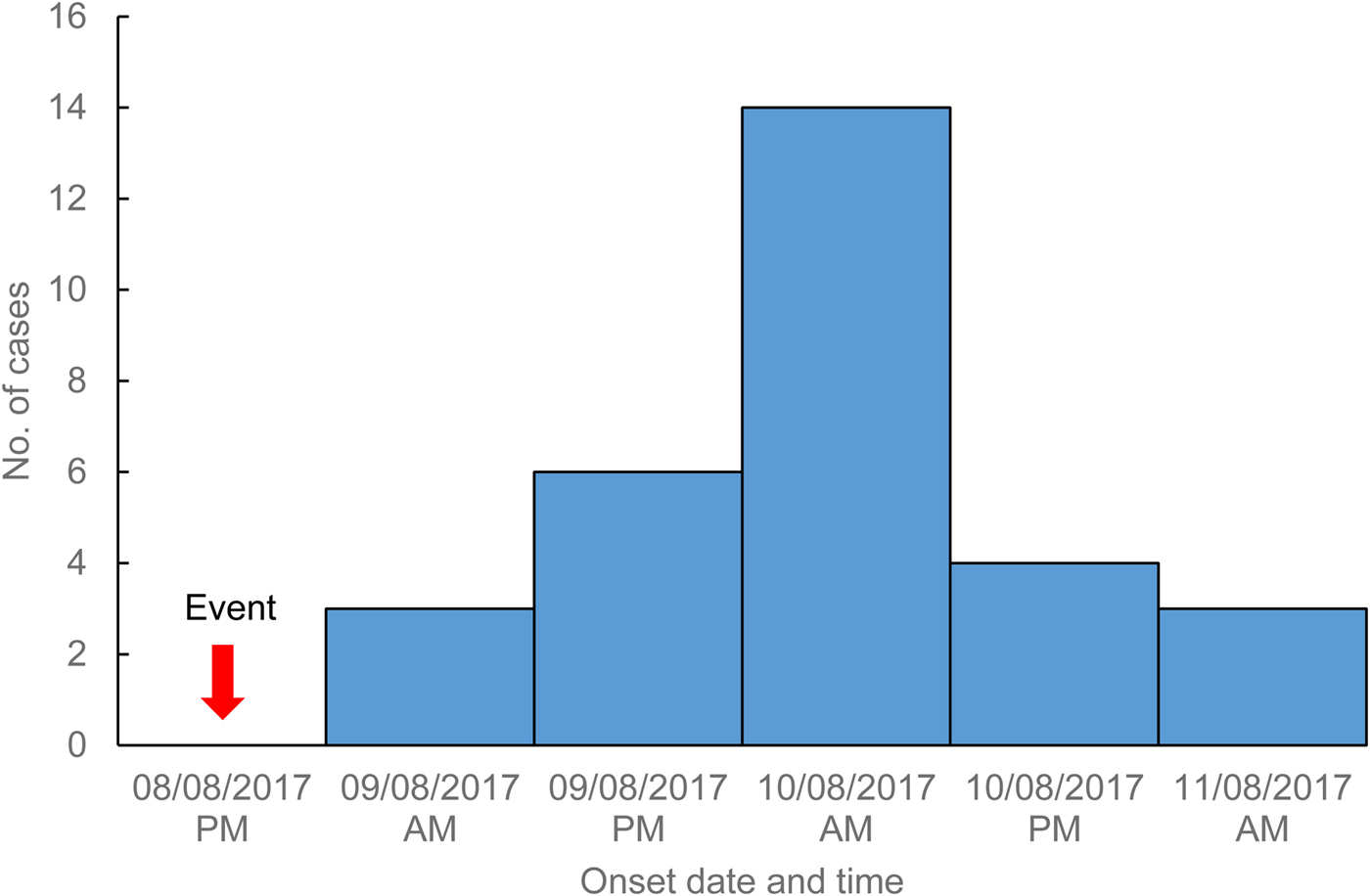
Fig. 1. Cases of gastroenteritis by illness onset date and time, Hunter New England, 9–11 August 2017.
The median age of cases was 48 years (range 28–70 years) and 75% (n = 24) were female, consistent with the demographic profile of all attendees. The case symptom profile was typical of norovirus with commonly reported symptoms including diarrhoea (n = 28, 88%), abdominal pain (n = 24, 75%), fatigue (n = 24, 75%), nausea (n = 23, 72%) and vomiting (n = 20, 63%). Less common symptoms included muscle aches (n = 15, 47%), fever (n = 13, 41%) and headache (n = 13, 41%). Three cases sought clinical care from a general practitioner, with a further two cases presenting to a hospital emergency department, with one requiring overnight admission.
Two stool specimens were submitted for testing, with positive reverse transcription polymerase chain reaction (RT–PCR) results for norovirus. RT–PCR targeting a 575 bp ORF1/ORF2 overlapping region was used for genotyping of GI and GII noroviruses [Reference Lim9]. Further genotyping identified a GII.P16/GII.4 Sydney 2012 norovirus.
A second questionnaire aimed at gathering further symptom details and information on secondary cases was sent to all cases (n = 32) 10 days after the first survey completion. Of the 32 cases, 23 (72%) completed the second questionnaire. The median duration of gastrointestinal symptoms (diarrhoea or vomiting) was 59 h (range 4 h–13 days). The case reporting the longest diarrhoea duration of 13 days was known to have an underlying medical condition and described a period of symptom resolution followed by a relapse of diarrhoea. Three cases (13%) reported secondary illness in four household contacts following the event at the time of the second questionnaire completion.
Investigation into the outbreak did not identify a definitive source or index case. The event had a catered sit-down menu with dishes served alternately. A univariable analysis of food consumed at the event identified that cases had significantly increased odds of having consumed ravioli with a risk ratio of 6.1 (95% CI 2.24–16.7, P < 0.001), with 83% of cases reporting eating the ravioli. Despite strong epidemiological evidence to suggest this dish as the transmission vehicle, a food safety inspection of the function premises did not identify any procedural or hygiene issues and there were no reports of sick food handlers. Seven attendees reported being in contact with individuals with similar symptoms in the week prior to the event, and three attendees developed symptoms prior to the minimum incubation period of 10 h [Reference Heymann1]. No cases reported symptoms of diarrhoea or vomiting during the event.
This case report describes an outbreak of a newly emerging strain of norovirus GII.P16/GII.4 Sydney 2012. Since late 2016, this new recombinant strain was also detected in several countries including France, South Korea, the UK and the USA [Reference Bidalot6, Reference Ruis8, Reference Choi10]. Genetic analysis of this genotype has suggested that changes in the GII.P16 RNA-dependent RNA polymerase in combination with predominant GII.4 capsid protein may provide this recombinant strain with increased transmissibility and a competitive advantage [Reference Ruis8, Reference Choi10].
Three of the five 2017 norovirus outbreaks from the Hunter New England region that had genotyping performed were found to be the new GII.P16/GII.4 Sydney 2012 genotype, suggesting it could be emerging as a predominant strain in this region. Further full-length sequencing and strain comparison has been undertaken by Lun et al. (GenBank accession number KY905335) [Reference Lun11]. The other two norovirus outbreaks from the Hunter New England 2017 winter season were identified as the common pandemic GII.Pe/GII.4 Sydney 2012 strain, and the newer GII.P16/GII.2 strain, which was associated with outbreaks in China and Germany in 2016–2017 [Reference Ao5, Reference Niendorf7].
Surveillance data on all institutional gastroenteritis outbreaks in the Hunter New England region has shown an increasing trend in the proportion of confirmed norovirus outbreaks in 2016 and 2017. Of the 120 institutional gastroenteritis outbreaks reported in 2017 14% were confirmed norovirus compared with 11% of the 62 outbreaks in 2015. The largest increase has been in gastroenteritis outbreaks of unknown aetiology (95 outbreaks in 2017 compared with 51 in 2015), the majority of which are presumably norovirus based on symptom profile, attack rate and transmission pattern. The increase noted in 2016–2017, coinciding with the emergence of the GII.16/GII.4 Sydney 2012 norovirus strain in NSW, is similar to what was observed in 2012 and 2013 following the emergence of the GII.Pe/GII.4 Sydney 2012 pandemic strain. Whilst the apparent increasing trend in institutional outbreaks may be associated with the emergence of the GII.P16/GII.4 Sydney 2012 strain, a more conclusive assessment of circulating norovirus strains is limited by the small number of outbreaks that have samples sent for testing and then further genotyping.
The Research Electronic Data Capture (REDCap) software used to develop and rapidly disseminate an event-specific questionnaire to function attendees on the day of outbreak notification allowed a large number attendees to be reached in a short period of time with the rapid collation of data that would be unachievable with conventional phone interviewing techniques [Reference Harris12]. Whilst there were some limitations to the use of an online questionnaire, such as the inability to clarify dates and time or further symptomatology, it proved to be a highly useful tool in this outbreak setting.
The GII.P16/GII.4 Sydney 2012 norovirus genotype appears to be emerging as a common strain in the Hunter New England region. Further surveillance and norovirus typing in NSW and across Australia would provide a more comprehensive picture of the extent and importance of this emerging strain in Australia and its potential to become the next pandemic strain.




