INTRODUCTION
Japanese encephalitis (JE) is an important arboviral infection of public health concern. Japanese encephalitis virus (JEV), a member of Flaviviridae, has established an intricate relationship among insect vectors, vertebrate reservoirs and human beings, where the latter acquire infection as accidental hosts [Reference Petersen and Marfin1].
JE presents commonly as an acute encephalitic syndrome, the ratio of clinical and subclinical infection being 1:300 to 1:1000. The case-fatality rate varies between 10% and 30% with residual neurological sequelae in one-third of survivors [Reference Solomon2].
The virus is maintained in nature as a single-stranded positive-sense RNA of ~11 kb in length. The open reading frame encodes for a polyprotein which in turn is spliced into seven non-structural proteins (NS1, 2A, 2B, 3, 4A, 4B, 5) [Reference Sumiyoshi3, Reference Chambers4] and three structural proteins (C, M, E). The E protein, which has three structural domains mediates viral attachment, entry, viral assembly and elicits potent neutralizing antibodies. Of the three structural domains, domain III contains the putative receptor-binding activity, domain II encodes the fusion loop involved in pH-dependent fusion of the virus with the host cell membranes, and domain I participates in the E-protein structural rearrangements required for fusion [Reference Solomon2, Reference Samuel and Diamond5]. Because of its putative role in neuroinvasiveness, this region is under constant immune selective pressure and provides sufficient information to infer evolutionary and epidemiological relationships [Reference Uchil and Satchidanandam6].
In recent years, apart from the rise in incidence of JE cases and associated deaths in endemic areas, the spread of this disease has also been observed in previously naive localities [Reference Hanna7]. The changing epidemiology along with the error-prone nature of the JE viral RNA-dependent RNA polymerase have lead to an increase in the diversity and neurovirulence of the virus. India has been experiencing a series of epidemics of JE since 1955 in various states with variable propensities. The state of Haryana in North India had its first episode of JE in 1990, since then similar epidemics have occurred at regular intervals in the rice-growing regions of the state [Reference Rao8]. The success of the JE control programme through live attenuated vaccine (SA-14-14-2) might depend on the molecular epidemiology of the virus genotypes circulating continuously in this region. In various animal experiments, single amino-acid substitutions in the E gene have been shown to alter the neuroinvasiveness of JE isolates [Reference Arroyo9–Reference Ni and Barrett12], necessitating the need for monitoring JEV strain variations at the molecular level in clinical isolates.
METHODS
Study subjects
A total of 95 patients with suspected viral encephalitis during October to December 2007 were enrolled in this study with their informed consent. These patients were from the village areas of Haryana, Uttar Pradesh and Punjab and were admitted to the in-patient departments of the Postgraduate Institute of Medical Education and Research (PGIMER), Chandigarh, India. One hundred and forty-two samples were collected from these 95 patients (paired CSF and blood in 47 patients, only CSF in 12 and only blood in 36 patients).
The samples were transported on ice to the Virology Laboratory as soon as possible after sample collection. Both the serum and CSF samples were aliquoted and stored at −70°C until further processing. Precautions were taken to prevent repeated freezing and thawing of the samples.
All the samples were subjected to detection of IgM antibody by JEV-specific MAC-ELISA (NIV, India) and viral RNA by JE specific RT–PCR. The JEV IgM antibody-negative samples were screened for West Nile virus (WNV) specific IgM antibodies using micro-ELISA (Panbio, Australia) to rule out the possibility of WNV infections.
Extraction of viral RNA
The JEV reference strain (GI no. AF080251, P20078; NIV, India) was grown in the Vero cell line. The successful propagation of the virus was confirmed by immunofluorescence and the culture supernatant was used as positive control in RT–PCR as previously described [Reference Swami13]. Viral RNA was extracted from 140 μl tissue culture lysate and clinical samples (CSF and serum) using QIAamp Viral RNA Mini kit (Qiagen GmbH, Germany), according to the manufacturer's instructions. The RNA extracted from the samples was directly digested on column with DNase I. Total RNA thus obtained was subjected to RT–PCR.
cDNA synthesis and E-gene amplification
The reverse transcription was performed in the presence of random hexamer primers. The oligonucleotides for nested PCR were selected from the conserved regions of the E protein genes, i.e. the outer primers, JEGAD3: 5′-AGG AAT CCT GGC TAT GCT TTC CTG-3′ (874–887) and JEC1: 5′-AGC ATG CAC ATT GGT CGC-3′ (2460–2477) and the inner primers, JEC4: 5′-GTG CAC ATG CCA TAG GT-3′ (1875–1892) and JER2: 5′-GTG CAT GGA ACC ACC ACT-3′ (1404–1421) [Reference Swami13, Reference Paranjpe and Banerjee14]. The amplification was performed in a final PCR reaction volume of 50 μl containing 5 μl c-DNA, 1× PCR buffer premixed with MgSO4, 200 μmol of each dNTP, 50 pmol each of forward primer set and 1·5 U of high-fidelity p.f.u. Taq polymerase (Promega, USA). Amplification started with an initial incubation at 95°C for 1·5 min to denature DNA and to activate DNA polymerase, primer annealing at 55°C for 1 min and extension for 3 min. It was followed by 30 amplification cycles consisting of denaturation at 94°C for 1 min, annealing at 58°C for 60 s and extension at 72°C for 30 s. Amplification ended with an extension step at 72°C for 10 min. For the second round of PCR 5 μl PCR-I amplicon was taken as the template and the inner set of primers was used, implementing the same protocol. Next, 1·5% (w/v) agarose gel electrophoresis (0·5 μg/ml containing ethidium bromide) was performed in order to visualize the 488-bp amplified product.
DNA sequencing
The nested primers, JEC4 and JER2, were used as forward and reverse primers, respectively, for sequence determination. The PCR products were purified with a QIAquick gel extraction kit (Qiagen, Germany) and the purified PCR products were cycle sequenced in the forward and the reverse direction in an ABI PRISM 310 genetic analyser (PE Applied Biosystems Inc., USA) by using an ABI PRISM Big Dye Terminator cycle sequencing ready reaction kit (PE Applied Biosystems Inc.). The nucleotide sequences were submitted to Genbank with the accession numbers GQ398493, GQ398494, GQ398495, GQ398496, GQ398497, GQ398498 and GQ398499.
Phylogenetic analysis and mutation detection
The nucleotide sequences of the E protein gene were aligned with the available corresponding JEV sequences from Genbank using CLUSTAL X version 2.0.10 [Reference Higgins, Thompson and Gibson15]. Genetic distances were calculated using the Kimura two-parameter matrix. Phylogenetic trees were constructed by the neighbour-joining method [Reference Saitou and Nei16]. To confirm the reliability of the pairwise comparison and phylogenetic tree analysis, bootstrap re-sampling was performed on 1000 datasets. Using MEGA (molecular evolutionary genetic analysis) version 4.0.2, phylogenetic and molecular evolutionary analyses were performed [Reference Kumar, Tamura and Nei17]. The E-gene sequences of the previous Indian isolates, JE isolates from Genbank and of our field strains were translated into protein sequences in silico for the detection of synonymous and non-synonymous mutations at positions 142–306 of the E protein [Reference Arroyo9, Reference Bikandi18]. The amino-acid positions 176, 177, 227, 244, 264 and 279 were focused as they have been observed to play a major role in the neuroinvasiveness as well as neurovirulence of the JEV isolates in animal experiments [Reference Arroyo9].
RESULTS
Twenty-three of the 95 patients showed the presence of JE-specific IgM antibodies either in CSF alone or in both CSF and blood. Thus, they were considered to be confirmed cases of JE. Fourteen patients had IgM positivity in serum alone and were considered as probable cases of JE [Reference Hills19]. JEV RNA was detected in the CSF samples of seven cases (30·4%, 7/23) by RT–PCR. None of the 58 JE-negative samples had IgM antibody against WNV.
Of the 23 confirmed cases, 70% belonged to the pediatric age group, with a male:female ratio of 3:1. Ninety percent of the patients with proven JE resided in rural areas. These patients presented with moderate to high-grade fever (41%), convulsions (65%), neck rigidity, posturing and abnormal movements (35%). Convulsion was often the presenting symptom. The serum biochemical parameters were within the normal limits. CSF cultures yielded no bacterial growth. A mortality of 27% was observed in JE-positive cases.
The E gene-amplified product of 488-nt sequence revealed 95% homology with the JEV genome from geographically diverse JEV isolates. The field strains exhibited a nucleotide sequence identity of 97% and 96% with the Nakayama strain and the first Indian isolate of JEV, respectively. The present isolates belonged to genotype III by phylogenetic analysis (Table 1, Fig. 1).
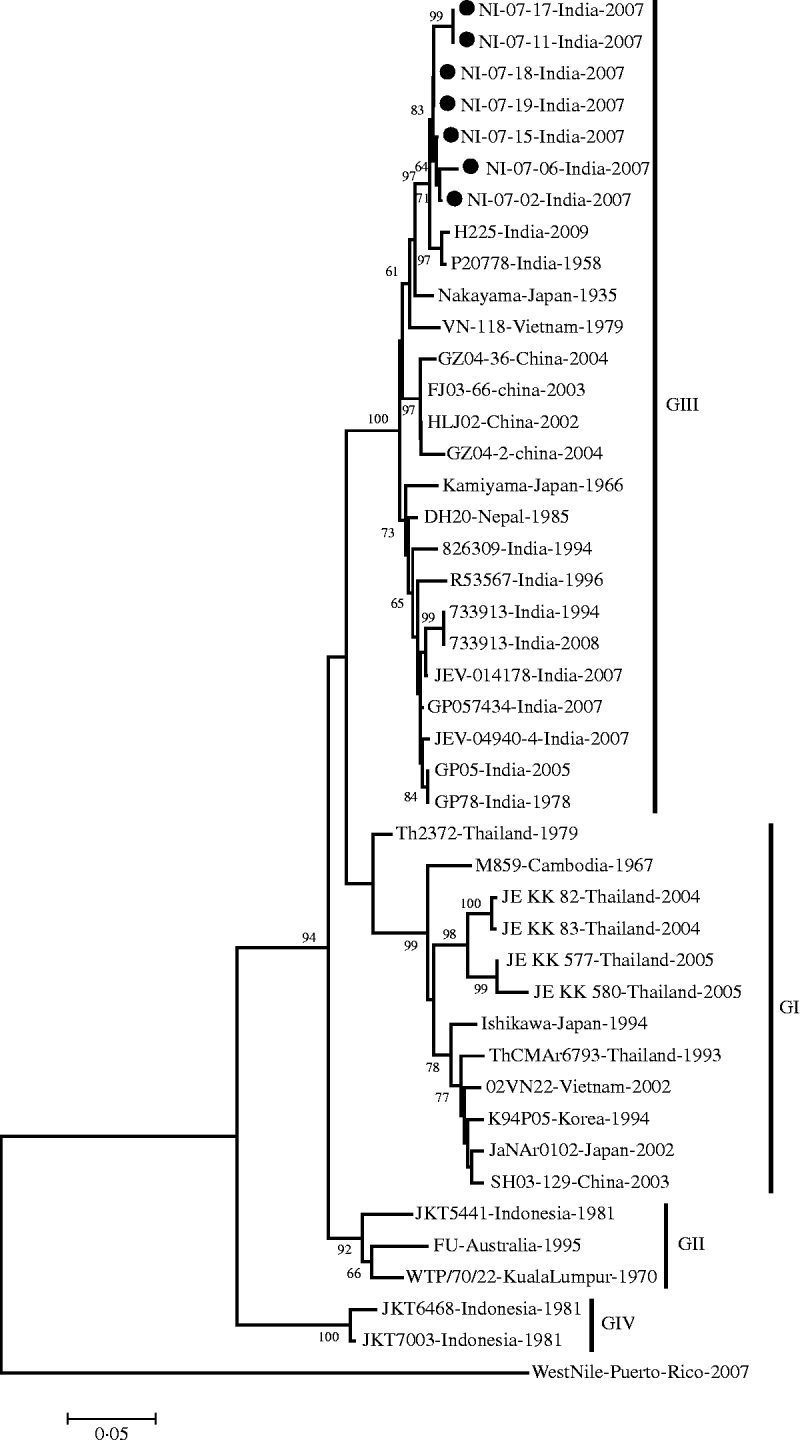
Fig. 1. Phylogenetic tree constructed by the neighbour-joining algorithm based on partial E-gene sequence of Japanese encephalitis virus isolates, with reference to other South East Asian isolates. Present field strains are marked with solid circles (•). West Nile virus E-gene sequence (gi|225690821) was used as outgroup. Bootstrap values are shown at major branch points. The genetic distance was calculated at the rate of 0·05.
Table 1. Strains of Japanese encephalitis virus used for phylogenetic analysis
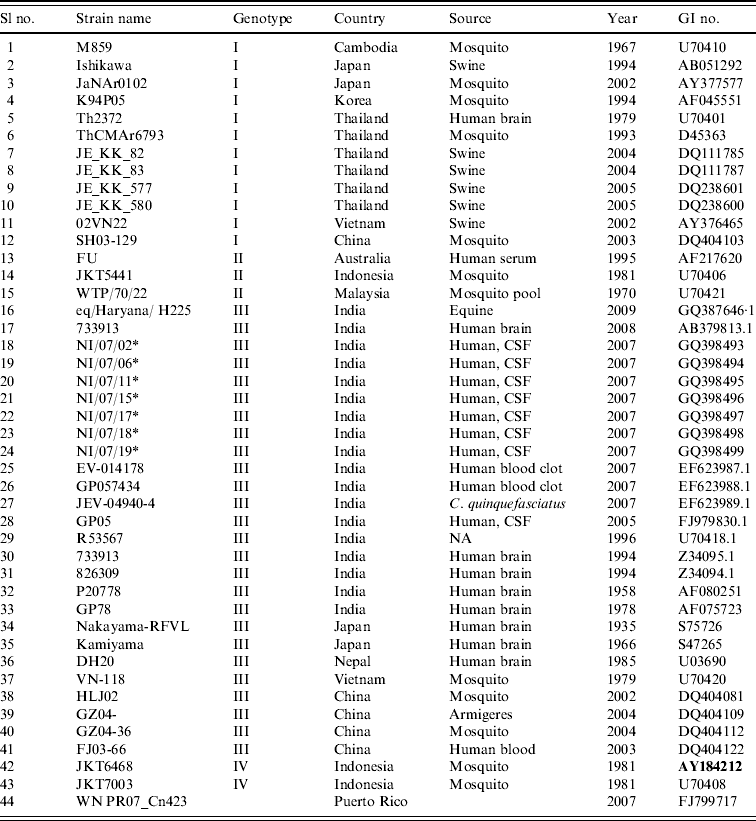
* Field strains.
On mutational analysis at the various positions of the E protein (Table 2, Fig. 2), the present JEV strains revealed a point mutation (S>T) at position 227 replacing serine (S) with threonine (T) compared to all other Indian isolates and other JEV reference strains.

Fig. 2. The amino-acid sequence (E protein) comparison between Indian isolates and other overseas strains.
Table 2. Variations of amino acids at important positions in the E protein of some representative strains

Bold indicates mutations detected in the study.
DISCUSSION
JE is an endemo-epidemic disease spreading over the agricultural regions of South East Asia. The high ambient temperature and waterlogged paddy fields, coupled with pig rearing and global climatic change, have facilitated JEV transmission.
The search for genetic determinants of virulence in animal models of flavivirus encephalitis has focused on the E protein [Reference McMinn20, Reference Heinz21]. This protein comprises 500 amino acids. The structural and functional integrity of this polypeptide relies on its three distinct domains. Domain III is projected over the viral outer membrane and acts as the receptor-binding domain with domain II being the dimerization domain. Domain I lies in between domains II and III and acts as a hinge region providing flexibility to the structural E protein (Fig. 3). Domain I consists of 127 residues (1–51, 135–193, 283–299); domain II has 172 residues (52–134, 194–282) and domain III is a contiguous stretch of 100 residues (300–399). It evokes neutralizing antibodies and protective immune responses in the host [Reference Heinz21, Reference Mason22]. Besides being a cell receptor-binding protein it also mediates membrane fusion and cell entry.

Fig. 3. Predicted three-dimensional structure of E protein of Japanese encephalitis virus (Indian strain 27 708) taking PDB ID 2HG0 as the template. The domains, locations and amino-acid positions 176, 177, 227, 244, 264, and 279 are shown.
The significance of point mutation has already been accepted during the recent resurgence of Chikungunya epidemic where a 226 A>V point mutation in the E1 gene played a key role in the adaptation of the virus to the Aedes albopictus vector, viral infectivity, and effective transmission [Reference Tsetsarkin23]. The three-dimensional structure of the E protein of the first Indian JEV isolate, Vellore 20778 strain was modelled taking the crystal structure of the closely related WNV as the reference template. Six amino-acid positions 176, 177, 227, 244, 264 and 279 were mapped on the three-dimensional structure of the E gene. These particular positions were targeted because of their contribution to the neurovirulent nature of JEV in animal models. Interestingly, the present isolate had a non-synonymous point mutation of S227T corresponding to the loop region in domain II of the E protein (Fig. 3). This molecular change at the amino-acid level has neither been observed in previous Indian isolates nor in isolates from other countries suggesting that the S227T amino-acid change is a novel mutation. There are several mutations which are involved in the pathogenesis of JEV. A mutation at the 138 amino-acid position (E protein, E>K) in JEV significantly altered the multiple steps of the viral life-cycle and even potentially induced substantial attenuation of JEV [Reference Zhao24]. In an independent study, Tajima et al. have shown that the S123R mutation in the E protein exhibited a significantly increased growth rate in N18 cells and virulence in mice [Reference Tajima25].
On the basis of the divergence of nucleotide sequences of capsid/premembrane protein (C/PrM) and envelope (E) genes, five different genotypes of JEV have been identified, where genotypes I–III (GI–III) are distributed throughout southern Asia; GIV strain was isolated from eastern Indonesia and an isolate originating in Malaysia represented the fifth genotype [Reference Uchil and Satchidanandam6]. The present JEV strains obtained in North India belong to genotype III in accord with the previously circulating JE strains in various parts of South East Asia [Reference Parida26–Reference Wang28].
Understanding the genetic changes in the envelope gene is an important step in understanding the evolution of JEV. Extensive surveillance is now essential in order to understand the geographic movement and genotype shift of JEV. The present mutation observed might turn out to be significant, particularly in terms of virulence and vector competency.
ACKNOWLEDGEMENTS
The authors acknowledge the financial grant received from the Indian Council of Medical Research, New Delhi for this work. We are grateful to Dr Eric Summer, programme director, ISID, USA and Dr Deepti Gupta, PU, Chandigarh for contributing towards the corrections in English language of the manuscript. S.K.P. is a recipient of a Senior Research Fellowship from the Council of Scientific and Industrial Research, Government of India.
DECLARATION OF INTEREST
None.







