Norovirus is estimated to cause 21 million US cases of acute gastroenteritis (AGE) annually and frequently causes AGE outbreaks in healthcare facilities, including long-term-care facilities (LTCFs) [Reference Scallan1]. Because of low infectious dose, environmental persistence, and prolonged symptomatic illness in hospitalized populations, outbreaks represent a substantial burden in patient morbidity and facility costs [Reference Lopman2, Reference Johnston3]. Clostridium difficile infection (CDI) is the leading US cause of healthcare-associated diarrhoea and acute diarrhoeal illness in LTCFs [Reference Simor4]. This disease burden is explained in part by the emergence and spread of a hypervirulent epidemic strain, BI/NAP1/027, which is associated with increased mortality, complications, and inadequate treatment responses [Reference Blossom and McDonald5].
Increases in norovirus outbreaks and rapid spread of CDI in LTCFs demonstrate co-infection is possible, although documented co-infection has rarely been reported [Reference Bignardi, Staples and Majmudar6]. We describe an AGE outbreak in a LTCF, including four cases of co-infection in order to better understand the epidemiology and interactions between these organisms.
During February and March 2012, an outbreak of AGE occurred at a 120-bed LTCF comprising three units (A, B, C) with 45, 45, and 30 beds, respectively. Cases were defined as AGE manifesting as ⩾1 episode of vomiting or diarrhoea (defined as ⩾2 loose stools within a 12-h period) among in-patients or staff who were present at any of the LTCF units during 1 February–15 March 2012. Patient chart reviews were conducted to obtain demographic and clinical information. To understand whether comorbidities differed in those affected by AGE, seven categories of chronic illness were considered: cardiovascular disease, diabetes, chronic kidney disease, gastrointestinal disease, neurological disease, pulmonary disease, and psychiatric disease. These categories were derived from review of the medical literature describing risk factors for and comorbidities associated with C. difficile and norovirus infection. Clinician medical record review using past medical history and current medication list classified chronic disease characteristics of the patients.
Case reports from occupational health records at the LTCF were used to identify cases in staff. Environmental testing for adenosine triphosphate bioluminescence, using Clean-Trace™ NGi Luminometer (3M™, USA), was performed in clinical and staff areas. This testing provides objective information on cleanliness of healthcare environments. High bioburden was defined as >1000 relative light units.
Because testing in this outbreak was initiated by clinicians, in the setting of medical decision-making, not all patients who were suspected cases were initially tested for both pathogens, or if C. difficile positive, for toxin production by the facility. Additionally, in accordance with the facility CDI diagnosis algorithm, the diagnosis of CDI did not require documentation of C. difficile toxin production. CDI in the facility diagnosis algorithm was defined as the presence of diarrhoea (defined as ⩾2 loose stools within a 24-h period) and a positive C. difficile toxin B gene polymerase chain reaction (PCR) test (personal conversation with facility infectious disease physicians and laboratory scientists). The facility diagnosis algorithm is based on the Infectious Disease Society of America and the Society for Healthcare Epidemiology of America clinical practice guidelines [Reference Cohen7]. Initial testing of stool samples for C. difficile was performed using multiplex real-time PCR (Cepheid Gene Xpert C. difficile PCR assay®, Cepheid, USA) to detect the toxin B gene (tcdB). Testing for norovirus was performed on stool samples from patients using reverse transcription (RT)–PCR, essentially as described previously [Reference Vega8]. Only the identified co-infected patients were further tested for C. difficile toxin (ImmunoCard® Toxins A & B, Meridian Bioscience, USA). C. difficile tcdC gene PCR and sequencing, and norovirus RT–PCR detection and capsid sequencing for genotype and outbreak strain identification were performed on co-infected cases as previously described [Reference Vega8–Reference Spigaglia and Mastrantonio10]. Staff stool samples and medical records were unavailable for analysis.
Thirteen patients (11 from unit A and one each from two adjoining units) experienced AGE symptoms during 1–7 February. Six patients experienced AGE on unit A before the appearance of the first patient with AGE symptoms on adjoining units. Simultaneously, four staff (three from unit A) with AGE were identified. During 1 February–15 March 2012, a total of 30 patients and 29 staff experienced AGE. Figure 1 illustrates the epidemic curve for the outbreak.

Fig. 1. Acute gastroenteritis (AGE) cases by date of symptom onset and unit location, February–March 2012.
The patient AGE attack rate varied by unit: 30·4% (21/69) on unit A, 8·2% (6/73) on unit B, and 5·7% (3/53) on unit C. Of the 29 affected staff, 28% (8/29) were assigned to unit A, 28% (8/39) to unit B and 10% (3/29) to unit C. The remaining staff were primarily stationed in other locations in the hospital but probably spent time on the identified units during the outbreak. In patients, the CDI attack rate was 13% (9/69) for unit A, 6% (3/73) for unit B and no CDI cases were identified on unit C. The norovirus specific attack rate was 13% (9/69) for unit A, 4% (2/73) for unit B and no confirmed norovirus cases were identified on unit C. Median patient age was 69 years (range 55–99 years) with a median of four chronic conditions (range 2–6).
Figure 2 illustrates details of patient and staff testing and results. Twenty-five patients were tested for both norovirus and C. difficile by PCR; four for C. difficile only (all negative); and one was not tested for either. Of 25 patients tested for both pathogens, seven (28%) were PCR-positive only for C. difficile; seven (28%) were PCR-positive only for norovirus; seven (28%) were negative for both pathogens; and four (16%) were positive for both pathogens. Two of the four co-infected patients tested positive for C. difficile toxin production. All four co-infected patients were determined to be clinically positive according to the facility's CDI definition described above. None of the seven patients PCR-positive only for C. difficile were tested for C. difficile toxin production. Norovirus capsid sequences indicated all four were identical genotype II.4 New Orleans strains, and all four C. difficile strains were identical 027 strains. Two stool samples from staff tested positive for norovirus by PCR at outside facilities; no additional testing of staff was identified by occupational health record review. All CDI patients received standard antibiotic treatment. Two deaths occurred, one norovirus-infected and one negative for both pathogens. AGE was not the primary cause of death in either patient. Two of the four co-infected patients had vomiting; none had fever. Three of the four co-infected patients resided in the same unit, and all had received antimicrobial therapy ⩽6 weeks before diagnosis. None had previously tested positive for CDI despite all having been tested previously. Table 1 describes the clinical characteristics of patients by infection status at symptom onset.
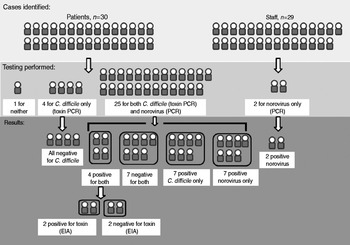
Fig. 2. Study population, testing and results. PCR, Polymerase chain reaction; EIA, enzyme immunoassay.
Table 1. Clinical characteristics of patients by infection status at symptom onset, n (%)

* Seven categories of chronic illness considered: cardiovascular disease, diabetes, chronic kidney disease, gastrointestinal disease, neurological disease, pulmonary disease, and psychiatric disease.
† Any change in C. difficile infection (CDI) medication from metronidazole to vancomycin during initial treatment course.
‡ Any recurrence of CDI symptoms within 8 weeks of follow-up or receipt of an additional course of antibiotics for CDI at least 2 weeks after completion and response to initial course.
§ Length of time from symptom onset to death was 79 days and 23 days, respectively.
¶ Resolution of diarrhoea and no need for further treatment.
∥ Tested for both pathogens.
Environmental testing revealed patient care and nursing areas with high residual bioburden, including nursing station keyboards, medication preparation carts, patient telephones, bedside rails, nursing conference room telephone, and white board markers. Additional observed infection control lapses included shared food and water left in patient care areas, limited enforcement of isolation and cohorting of patients, and inadequate surface cleaning.
No source of the norovirus outbreak was identified although it seems likely to have originated from staff or visitors to unit A given the attack rate and appearance of initial symptomatic patients at this location. In addition, unit A is a subacute ward with more acute patients compared to patients on other units. Because these patients are often receiving additional services (dialysis, respiratory therapy, etc.) there is more movement of staff and patients. This level of care also generally has a higher staff to patient ratio. Both of these factors probably increased the risk of infection and transmission on unit A. Unit C is a hospice unit with minimal movement of patients and infrequent substitution of staff likely to limit the impact of the outbreak on this unit. Similarly, unit B is a stroke rehabilitation unit with less mobile patients. These factors probably explain the differences between attack rates by unit.
We identified a subset of AGE outbreak patients with C. difficile and norovirus co-infection during a simultaneous outbreak in a LTCF. Two of the four co-infected cases had evidence of C. difficile toxin production concurrent with norovirus infection. Clinical interactions between these pathogens have not been extensively studied, and whether pathological changes or changes in the lower intestinal microbiota associated with one infection increase the risk for acquiring the other is unknown. The increased need for isolation of patients during a norovirus outbreak may limit resources available for the isolation of patients with CDI, increasing the risk for C. difficile transmission. In addition, norovirus-induced diarrhoea during an outbreak may increase the dissemination of spores from patients who were previously asymptomatically colonized by C. difficile. Areas identified with increased residual bioburden suggest inadequate environmental cleaning and could increase the risk of disease transmission.
In LTCFs, CDI and norovirus outbreaks are common and can present a diagnostic dilemma. Increased C. difficile testing in LTCFs where frequent asymptomatic colonization with C. difficile occurs following recent antibiotic exposure can result in the misdiagnosis of CDI in patients with AGE due to other causes or a false association of CDI with norovirus AGE [Reference Wilcox and Fawley11]. Similarly, the diagnosis of norovirus is not always straightforward because certain patients asymptomatically shed norovirus and older persons (the majority of residents in LTCFs) in particular might have atypical presentations, including no vomiting or prolonged course, causing confusion with CDI [Reference Lopman2]. In this outbreak, only half of co-infected patients had vomiting suggesting the need for clinical vigilance during an AGE outbreak to identify and respond to suspected norovirus infection. At the same time, half of the co-infected patients had evidence of toxin production suggesting CDI algorithms including treatment should continue to be followed during a confirmed AGE outbreak as both organisms can present similarly in this population. It could be helpful for facilities to develop an AGE outbreak test panel to assist clinicians in performing appropriate testing in every suspected case.
Certain patients in this outbreak were not tested for both pathogens, resulting in possible underestimation of infection. Determining whether positive C. difficile PCR represented colonization or true co-infection is difficult because only two of four co-infected patients had evidence of toxin. It is the standard of the facility to use PCR for detection of toxigenic C. difficile as their CDI diagnosis tool. We did not have access to all samples from identified cases and were only able to test the four identified co-infected patients for toxin production. Because of this we were unable to compare the rate of toxin production positivity between co-infected patients and C. difficile-only infected patients. In addition, even though the manufacturer reported that the sensitivity of the toxin assay used in our investigation was 95% (Meridian Biosciences), presence of toxin may have been missed despite PCR evidence of toxigenic C. difficile. Even the detection of measurable toxin does not prove an aetiological role for C. difficile in a patient's diarrhoea as, for example, patients may remain toxin-positive for a period of time following successful treatment of CDI. Because this outbreak was limited to a single facility with relatively few cases, the prevalence of co-infection might not be generalizable to AGE outbreaks in other LTCFs. During concurrent outbreaks, more complete investigation of epidemiological linkage between infected patients and staff, and more complete laboratory testing, including C. difficile toxin testing could better describe the clinical relationship between these organisms and the impact of one on the rate of infection by the other.
Coexistence of both organisms and overlapping presentations has practical implications for treatment and diagnosis of CDI. During an identified norovirus outbreak in settings where C. difficile is prevalent, CDI testing of patients with acute diarrhoea should be considered because differentiation between organisms can be difficult on the basis of clinical presentation alone. C. difficile PCR-positive patients in norovirus outbreak settings should receive C. difficile treatment as it is possible that the positive test represents true infection and not simply colonization in the setting of a norovirus infection (as was suggested by toxin-positive co-infected cases). Further investigation into improved diagnostic choices in cases of possible co-infection would be helpful. For example, a toxin production test might be helpful in clarifying the specific cause of a patient's diarrhoea during an outbreak with both organisms. Better characterization of simultaneous outbreaks and co-infection in patients might assist in more clearly defining testing and treatment protocols in these settings.
ACKNOWLEDGEMENTS
The authors thank the patients and staff of the Community Living Center, Jane Inaura, NP, of Veterans Affairs Northern California Health Care System and the infection control staff who greatly assisted with the investigation. This investigation was supported by intramural funding from the Department of Veterans Affairs. The findings and conclusions in this report are those of the author(s) and do not necessarily represent the official position of the Centers for Disease Control and Prevention or the Department of Veterans Affairs, Health and Human Services or the U.S. Government nor does mention of trade names, commercial products or organizations imply endorsement by the U.S. Government.
DECLARATION OF INTEREST
None.





