INTRODUCTION
Tuberculosis (TB) is one of the most widespread infectious diseases and a leading cause of death for adults worldwide [Reference Cosivi1]. Although much attention has focused on treatment and prevention of human TB caused by Mycobacterium tuberculosis, zoonotic TB due to Mycobacterium bovis, known as bovine tuberculosis (bTB), has become an important re-emerging public health concern in developing countries. A large number of livestock keepers (pastoralists) combined with poor public health infrastructure, limited bTB control measures for cattle and animal products, and a large immunocompromised human population due to HIV/AIDS make Africa particularly vulnerable to the health impacts of bTB [Reference Etter2]. Zoonotic bTB infection is significant, as M. bovis is naturally resistant to pyrazinamide, a first-line TB treatment drug [Reference Cosivi1]. Both of these TB-complex mycobacteria have demonstrated the ability to move between animal (including wildlife) and human populations [Reference Alexander3, Reference Michel, Gibbs and Bokma4].
Bovine TB has also emerged as a disease of concern for wildlife conservation. Spillover from bTB-infected cattle herds was thought to have resulted in bTB infections in African buffalo (Syncerus caffer) inhabiting the southern portion of Kruger National Park (KNP), South Africa (Kloeck, 1998 cited in [Reference Michel5]). Within 15 years of the discovery of the first cases in KNP in 1990, bTB had spread through most of park's buffalo herds [Reference Rodwell6], and infections were documented in 10 other species [Reference Michel5]. Reported impacts of bTB in KNP and other South African parks included decreased body condition and drought tolerance of infected buffalo [Reference Caron, Cross and du Toit7], lowered buffalo reproductive success [Reference Jolles, Cooper and Levin8], and mortality and disruption of pride dynamics in infected lion prides [Reference Michel5]. In East Africa, bTB infections have been documented since the 1960s in buffalo in Uganda [Reference Woodford9, Reference Kalema-Zikusoka10] and recently in wildebeest (Connochaetes taurinus), topi (Damaliscus lunatus), and one lesser kudu (Tragelaphus imberbis) in northern Tanzania [Reference Cleaveland11]. However, little is known about whether or not bTB is present in other ecosystems, and population-level impacts of bTB have not been characterized.
The Ruaha ecosystem of south-central Tanzania encompasses vast wildlife protected areas of high conservation value bordered by lands inhabited by large numbers of livestock. Farmers, traditional livestock keepers, livestock, and wildlife inhabiting the southern portion of the Ruaha ecosystem all depend upon water from the Great Ruaha River and its tributaries (Fig. 1). Over time, human migration due to protected area creation and other government resettlement programmes [Reference Williams12] coupled with the seasonal drying of the Great Ruaha River largely due to upstream diversion for agriculture [Reference Lankford13] has altered the distribution of people, livestock, and wildlife. Heightened competition for land and water resources has increased wildlife conflict and concerns that increased overlap among human, wildlife, and livestock populations in Ruaha may be increasing the risk of zoonotic disease transmission including bTB [Reference Franks, Lankford and Mdemu14, Reference Mazet15].
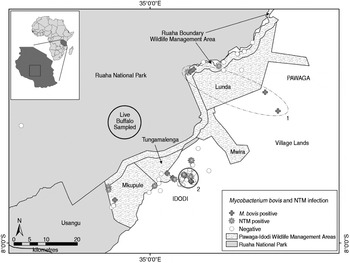
Fig. 1. Spatial distribution of M. bovis and non-tuberculosis complex mycobacteria (NTM) infected and uninfected wildlife carcasses (n = 121), and the approximate sampling area where 30 live buffalo were tested for bovine tuberculosis in a livestock–wildlife interface area in the southern portion of the Ruaha ecosystem, south-central Tanzania. Although not statistically significant, an elliptical (no. 1, P = 0·057) and circular (no. 2, P = 0·066) shaped region of a higher than expected number of M. bovis cases (nos. 1 and 2), as indicated by spatial scan statistics are shown. The geopolitical division of village lands (Idodi or Pawaga) is shown in capital letters.
Despite data indicating widespread bTB infection in cattle surrounding Ruaha's wildlife protected areas, it was not known if wildlife in the Ruaha ecosystem had been infected. Prevalence of bTB in cattle grazing the southeastern portion of the Ruaha ecosystem was estimated to be 13%, with 51% of sampled herds containing one or more positive reactors [Reference Kazwala16]. Previous wildlife data from Ruaha were limited to results from a sample of six wild animals [five elands (Taurotragus oryx) and one roan antelope (Hippotragus equinus)] that were serologically negative by enzyme immunoassay [Reference Cleaveland11]. Accordingly, we set out to determine if bTB was present in Ruaha's wildlife as part of a large-scale project assessing the impact of zoonotic diseases at the rapidly changing environmental interface of human, livestock, and wildlife populations in the Ruaha ecosystem.
We hypothesized that given widespread bTB presence in cattle, bTB would also be present in wildlife species inhabiting the livestock–wildlife interface areas south of Ruaha National Park (RNP) and that wildlife species would be infected with the same strain of M. bovis found in local cattle. We also examined the species and spatial distribution of M. bovis wildlife infections to determine potential for pathogen persistence in wildlife maintenance hosts, and identify potential high-risk areas for transmission that could be targeted as part of an ecosystem-based approach to reduce transmission of disease at the livestock–wildlife interface and improve animal and human health.
METHODS
Study area
The southern extent of the Ruaha ecosystem lies in northern Iringa District, south-central Tanzania (07°19′S to 07°36′S and from 35°05′E to 35°29′E). It covers about 30 000 km2 of different rangeland-use areas, RNP, the Rungwa, Kisigio and Muhesi game reserves, the Lunda-Mkwambi Game Controlled Area, the recently formed community-based Pawaga–Idodi Wildlife Management Area (PIWMA), and village lands. The area is internationally significant in terms of biodiversity conservation because it contains the only protected area system covering the transition between the vegetation communities of the Sudanian Acacia-Commiphora zone of East Africa and the Brachystegia (miombo) woodlands of southern Africa [Reference Williams12]. Our study was concentrated in the southernmost portion of the Ruaha ecosystem, comprised of rural villages, the PIWMA, and RNP (Fig. 1). Habitat consisted of patchily distributed semi-arid woodland and brushland, and active and fallow agriculture fields. Lands bordering the wildlife protected areas are heavily grazed, as evidenced by denuded vegetation, bare patches of soil, and the presence of many livestock and livestock faeces.
Ethics statement
All research activities in Tanzania were reviewed, approved, and permitted by the Tanzania Commission on Science and Technology (COSTECH), the Tanzania Wildlife Research Institute (TAWIRI), Tanzania National Parks (TANAPA), and University of California Davis Institutional Animal Care and Use Protocols nos. 12394 and 15919. No animals were killed for the purposes of this study.
Animal sampling
From 2006–2010, lungs, mediastinal and mesenteric lymph nodes, and other tissue samples from hunter-killed wildlife, opportunistically found carcasses, and wildlife depredated for causing crop damage in and around the PIWMA and RNP were collected by community game scouts trained to safely collect specimens, project veterinarians (D. Clifford, H. Sadiki), or technicians. The date, sex, age, and species were recorded for each animal sampled. Whenever possible the location of the carcass (latitude and longitude in decimal degrees, World Geographic System 1984 datum) was recorded using a handheld global positioning system (GPS) unit. For carcasses where GPS locations were not recorded, the nearest village or a locally known place name within the protected area was recorded by the scout. Collected tissues were examined for gross lesions and subsectioned: the outer surface of the tissue was seared, and then a sample for culture was collected using forceps that were chemically disinfected and a new sterile disposable scalpel blade for each animal. Specimens for culture were placed into sterile Whirlpak® plastic collection bags (Nasco, USA). Samples were frozen on average within 24 h of collection and then stored frozen at −20°C until testing.
Additionally, we received heparinized blood samples from 30 live African buffalo immobilized in RNP in 2011 during foot-and-mouth disease (FMD) surveillance efforts conducted by the South African Development Community and the Tanzanian Ministry for Livestock. Buffalo belonging to the Msembe herd that ranges in the southern portion of RNP were opportunistically immobilized via dart gun delivered from a helicopter. Additional buffalo herds inhabiting other areas of RNP were not sampled. Only buffalo aged 2–4 years were captured and sampled as they were the target age group for FMD testing.
Diagnostic testing
Culture and molecular diagnostics – wildlife tissues
Frozen tissues were thawed to room temperature, then pooled lung and lymphoid tissues were homogenized, decontaminated, and neutralized using standard methods [Reference Watt, Rayner and Harris17]. Resulting sediments were inoculated onto Lowenstein–Jensen media with pyruvate and Lowenstein–Jensen media with glycerol and incubated at 37°C for up to 12 weeks. Positive cultures with appropriate colony morphology [Reference Vestal and Kubica18] were subcultured onto another set of the same media for 3–4 weeks to obtain pure culture and then examined by microscope for the presence of acid-fast-bacilli (AFB) using Ziehl−Neelsen stain.
Heat-killed AFB-positive samples were further characterized by multiplex polymerase chain reaction (PCR), using primers to the 16S rRNA gene specific for the Mycobacterium genus and able to distinguish between M. avium and M. intracellulare, and primers aimed at the MPB70 gene of M. tuberculosis complex (MTC) organisms [Reference Wilton and Cousins19]. Samples with an amplification product of 1030 bp indicative of the genus Mycobacterium and of 372 bp were considered positive signals for MTC.
Spoligotyping was used to delineate the Mycobacterium species for animals with MTC and distinguish unique strain types. Briefly, PCR products representing all the spacer sequences in an isolate's genome were amplified using primers specific for the direct-repeat sequences of the direct repeat locus of the MTC chromosome. Hybridization of the amplification product mixture against a membrane to which the 43 individual spacer sequences were covalently linked was used to generate a specific pattern of positive and negative hybridization signals [Reference Kamerbeek20]. The absence of spacers 3, 9, 16, and 39–43 was used to classify isolates as M. bovis [Reference Kamerbeek20, Reference Costello21]. The M. bovis spoligotype patterns from wildlife samples were coded [Reference Dale22] and compared to spoligotypes SB0133 and SB0425 which were dominant in the cattle isolates from a study in an adjacent area in Mbeya and Iringa regions [Reference Kazwala23], and in 2007 from a cow from a village in Pawaga Division on the southern border of the PIWMA and RNP (Fig. 1).
Serology – live buffalo samples
Blood samples from live-captured buffalo were tested for bTB infection using a commercially available M. bovis gamma interferon (INF-γ) test kit, that utilizes a monoclonal antibody-based sandwich enzyme immunoassay to detect the production of INF-γ (Bovigam®, Prionics, Switzerland). Within 8 h of collection, heparinized blood samples from each animal were subdivided into three aliquots that were mixed with phosphate-buffered saline (nil antigen), bovine and avian purified protein derivative (tuberculin; Veterinary Laboratory Agency Weybridge, UK), respectively, and then incubated for approximately 20 h at 37°C. After incubation, plasma was harvested by centrifugation, frozen at −20°C, and transported to Sokoine University of Agriculture where the Bovigam assay was performed according to the manufacturer's instructions. All samples were tested in duplicate and positive and negative controls supplied by the manufacturer included on each 96-well plate. Animals were classified as positive bTB reactors if ODbovine − ODcontrol was ⩾0·049 and if ODbovine was greater than ODavian according to Whipple et al. [Reference Whipple24], with additional consideration given to samples with ODbovine ⩾0·385, as this value optimized test predictive value for sampled buffalo in South Africa [Reference Michel25].
Data analysis
Field and laboratory data were entered into an electronic database (Microsoft Excel, USA). The proportion of AFB-positive, M. bovis-infected, and non-TB complex mycobacteria (NTM) infected animals was calculated for all animals tested and for each species. Associations between M. bovis and NTM infection proportions, species group (defined as hoofstock, carnivores, primates, or small mammals), age (young vs. adult), sex, and sampling location (inside vs. outside of a wildlife protected area) were examined using Fisher's exact tests using Stata version 11.2 software (StataCorp., USA).
For carcasses that had known locations, a spatial scan statistic was used to determine if M. bovis infections were distributed randomly over space in our sampling area, and if not, to evaluate any spatial infection clusters for statistical significance (SaTScan version 9.1.1, M. Kulldorff, Harvard Medical School, Boston, MA, USA) [Reference Kulldorff and Nagarwalla26]. A Bernoulli model utilizing case-control (0/1) data was used, as it does not assume a homogenous distribution of the underlying population. A circular or elliptically shaped moving window was used to scan for spatial clusters encompassing from zero to not more than 50% of the locations by comparing the observed infections inside vs. outside the window using likelihood functions [Reference Kulldorff27]. Both the circular and elliptical window options were evaluated because the elliptical window allows for a better fit to linear geographical features, including roads and rivers, which may be associated with the location of carcass recovery due to ease of access [Reference Kulldorff28]. Maximum-likelihood estimates were generated by Monte Carlo simulations of 999 iterations and clusters evaluated for significance with P = 0·05. Spatial scan statistics were not used to evaluate clusters of NTM infections as location data was not known for all NTM-infected individuals.
RESULTS
Culture and molecular diagnostics – wildlife tissues
Tissue samples from 149 animals comprising at least 30 different species were collected. The exact number of species represented in the sample was not known as the specific species was not identified for three squirrels and 10/11 sampled mongooses. Kirk's dik-dik (Madoqua kirkii) and impala (Aepyceros melampus) were the two most commonly sampled species, comprising 36% of the total sample (Table 1). Hoofstock species comprised 59% of the sample (n = 88), with the remainder of sampled animals comprised of 24% carnivores (n = 36), 12% primates (n = 18), 4% small mammals (n = 6) and a single elephant. Adult animals were most commonly sampled (n = 125, 84%); 15 young animals were sampled, and age was not recorded for nine animals. Male animals comprised 70% of the animals sampled (n = 105); 33 females and 11 animals not having sex identified comprised the remainder of the sample. Variable numbers of samples were collected each year [2006 (n = 19); 2007 (n = 20), 2008 (n = 6), 2009 (n = 96), 2010 (n = 8)]. Of 124 animals whose exact or approximate locations were known, 38 (31%) were located within a wildlife protected area [PIWMA (n = 37), RNP (n = 1)]; while the remaining 86 individuals were located outside protected areas on village lands. Hoofstock were more likely to be sampled within protected areas than carnivores (one-sided Fisher's exact P = 0·048) and primates (one-sided Fisher's exact P = 0·009). All small mammals were sampled outside of protected areas.
Table 1. Results of acid-fast bacilli culture and mycobacterium PCR for 149 wild animals sampled from 2006 to 2010 in a livestock–wildlife interface area in and around the Pawaga–Idodi Wildlife Management Area and Ruaha National Park, Iringa Region, south-central Tanzania

AFB, Acid-fast bacilli; NTM, non-tuberculosis complex mycobacteria; MTC, Mycobacterium tuberculosis complex.
The number of animals positive/total tested (% positive) for acid-fast bacilli growth on culture, and tuberculosis and NTM is reported. All tuberculosis complex mycobacteria isolated were classified as Mycobacterium bovis by spoligotyping.
Cultures from 34 animals had positive AFB growth and were further characterized using the mycogenus PCR (Table 1). Of the 27 samples that produced amplification products, 12 belonged to the MTC, and 15 were NTM. M. avium and M. intracellulare were not identified. Yellow, white, or grey nodules were noted on gross examination of tissues from eight animals, but only one animal with gross lesions, an African lion (Panthera leo), had AFB growth on culture (Table 1).
Spoligotyping allowed classification of all 12 MTC isolates as M. bovis as they lacked spacers 3–7, 9, 16, and 39–43 (Table 2) [Reference Kamerbeek20, Reference Costello21]. M. bovis was isolated from 8% (12/149) of individuals sampled comprising eight species (Table 1). All 12 M. bovis isolates were spoligotype pattern SB0133 (Table 2), a previously recognized pattern in the M. bovis online spoligotype database belonging to the African 2 clonal complex of M. bovis [Reference Berg29]. The spoligotype pattern of wildlife samples was the same as those previously isolated from cattle sampled in Iringa and adjacent Mbeya region from 1993 to 1995 [Reference Kazwala23], and the single positive cow from our study area that was slaughtered and sampled in 2007 (Table 2).
Table 2. Spoligotype pattern (SB0133; www.mbovis.org) of 12 Mycobacterium bovis isolates obtained from a sample of 149 wild animals and a domestic cow inhabiting a livestock–wildlife interface area in the southern portion of the Ruaha ecosystem, south-central Tanzania

The approximate location of each case; Pawaga–Idodi Wildlife Management Area (PIWMA) or village lands (Village), and the nearest place name (in parentheses) with reference to Figure 1 is specified.
Although M. bovis infection was documented in 17% (3/18) of sampled primates, the primate infection proportion was not significantly greater than that of sampled hoofstock (8%, 7/88, one-sided Fisher's exact P = 0·227), or carnivores (6%, 2/36, one-sided Fisher's exact P = 0·200). M. bovis infection proportions were equivalent between sampled males (9%) and females (9%, Fisher's exact P = 1·00) and there was no difference in M. bovis infection proportions between animals sampled inside (8%, 3/38) and outside (10%, 9/86) wildlife protected areas (Fisher's exact P = 0·754). Infection with M. bovis was only detected in adult animals.
NTM infection was detected in 10% (15/149) of sampled animals belong to 11 species (Table 1). There was no difference in NTM infection proportion in species groups: 17% (1/6) of small mammals, 14% (5/36) of carnivores, and 10% (9/88) of hoofstock sampled were infected. No NTM infections were documented in sampled primates. NTM infection proportions were similar between sampled males (10%) and females (12%, Fisher's exact P = 0·758), and between sampled adults (12%) and young animals (15%, Fisher's exact P = 0·677). There was also no difference in NTM infection proportion between animals sampled inside (16%, 6/38) and outside (12%, 7/86) protected areas (Fisher's exact P = 0·215).
Exact locations for 12 M. bovis-infected animals and 109 uninfected animals were available for spatial analyses. A geographical cluster of four M. bovis-infected animals was detected inside a 143 km2 elliptical area that extended along the road from grazing lands through the Lunda portion of the PIWMA and up to the PIWMA/RNP border (no. 1, Fig. 1), but was not statistically significant (P = 0·057). A second smaller 10·9 km2 circular geographical cluster of three M. bovis-infected animals was detected in village lands (no. 2, Fig. 1), but was also not statistically significant (P = 0·066). Although neither cluster was statistically significant, less than one infected animal would be expected to have occurred in each of these areas if M. bovis infections were randomly distributed throughout the area where carcasses were found.
Serology – live-sampled buffalo
All 30 sampled buffalo were noted to be apparently healthy at the time of capture. Three buffalo (10%, 2 male, 1 female) were positive reactors according to test criteria established by Whipple et al. [Reference Whipple24]. Two of three positive reactors also had mean ODbovine values ⩾0·385 and thus would be considered positive using an alternate cut-off designed to maximize test predictive value in African buffalo developed by Michel et al. [Reference Michel25].
DISCUSSION
We document that bTB is present in wild animals inhabiting protected areas and village lands in the Ruaha ecosystem of south-central Tanzania. Our findings demonstrating M. bovis infection in 8% of sampled wild animals are consistent with findings from northern Tanzania [Reference Cleaveland11]. However, the opportunistic nature of the sampling effort in this study limits our ability to estimate the true prevalence of bTB in this population. Although weaker animals may be more likely to be successfully hunted, opportunistically found, or depredated, and thus potentially more likely to be infected than the general population, this potential bias in our data may be balanced by the fact that infected wildlife sampled in this study lacked clinical bTB lesions, and that trophy hunters prefer to select larger healthy males.
An unexpected finding was the documentation of bTB infection in eight different species occupying different ecological niches. To our knowledge, this is the first published report of M. bovis infection in free-ranging vervet monkey, yellow baboon, and Kirk's dik-dik in Africa, and the first published isolation of M. bovis in African buffalo and impala in Tanzania. M. bovis infection has been documented previously in chacma baboons (Papio ursinus) in South Africa [Reference Michel5] and in olive baboons (Papio anubis) from Kenya that were feeding on slaughterhouse offal from M. bovis-infected cows [Reference Tarara30, Reference Sapolsky and Else31]. Neither of these baboon species is present in the Ruaha ecosystem, but the yellow baboon fills a similar niche, being abundant both in protected areas and in village lands where they are adept at utilizing anthropogenic food sources. No published record of M. bovis infection was found for vervet monkeys, but M. tuberculosis infection has been reported in vervet monkeys held at a wildlife rehabilitation centre in South Africa [Reference Michel32].
Spoligotype similarity between wildlife sampled in this study and local livestock supports the hypothesis that livestock and wildlife are sharing pathogens. Infection of small carnivores and primates may indicate more recent spillover transmission from cattle as these species often live close to human settlements, have limited potential to maintain bTB in the absence of an alternate infection source (i.e. dead-end hosts), and often have short duration of illness [Reference Renwick, White and Bengis33]. The infected mongoose, blotched genet, vervet monkeys, yellow baboon, and Kirk's dik-dik in this study were all sampled near villages.
In addition to spillover, documentation of M. bovis infection in three species of large bodied, long-lived, gregarious herbivores (African buffalo, lesser kudu, impala) emphasizes the possibility for pathogen persistence in one or more wildlife maintenance hosts. Buffalo are the major wildlife bTB maintenance host in Africa, transmitting bTB within their herds without repeated spillover from cattle and serving as a source of bTB to other sensitive species, especially large carnivores [Reference De34]. Although there is no evidence indicating lesser kudu and impala are maintenance hosts, greater kudu with advanced bTB can shed large numbers of bacteria and may be able to maintain a separate infection cycle, as documented in KNP [Reference Renwick, White and Bengis33]. Once established, bTB infection in wildlife maintenance hosts can be a source of infection for livestock, thereby complicating control efforts or resulting in the re-emergence of bTB in livestock populations where the disease was formerly eradicated [Reference Quinn, Collins, Thoen, Steele and Gilsdorf35, Reference Schmitt36].
Our documentation of infected buffaloes in the community-based wildlife management area and in RNP has conservation and economic significance. Large herds of healthy buffalo attract revenue from both non-consumptive tourists and hunters coming to the game reserves surrounding RNP. Seasonal drying of the Great Ruaha River may reduce the spatial distribution of buffalo and cattle, compressing herds into a smaller area where potential for disease transmission is higher and forage competition is more severe. In addition, forage limitation, due to increased frequency and severity of bush fires, may be an ecological stressor that acts synergistically to cause disease (P. Coppolillo, unpublished data).
We did not identify a unique buffalo-only strain of M. bovis in Ruaha's buffalo, thus it is possible that the infections found were solely due to spillover from livestock. However, the fact that 3/30 young buffalo from a single herd sampled inside RNP were serological reactors strongly suggests that bTB is being transmitted buffalo-to-buffalo. Given the continued ecological threats, a wider systematic buffalo health assessment of multiple herds and age groups, coupled with updated herd demographic and spatial distribution data is needed to estimate the population prevalence of bTB, determine if bTB is widespread in buffalo herds in Ruaha, and determine if bTB or other disease could be contributing to any spatial range contraction or demographic changes.
In addition to conservation impact, bTB infection in wildlife has implications for human health. Buffalo, kudu, and impala are important hunted species, and dik-dik are commonly consumed bushmeat. Although the risk of contracting bTB from consumption of well-cooked meat is minimal for most people, organ meat (including lungs) is commonly consumed and most carcasses are processed in the field with little or no sanitary precautions. These factors coupled with a relatively high prevalence of infection with HIV/AIDS may put people processing and consuming hunted wildlife at greater risk of contracting and developing clinical TB [Reference Cosivi1].
Implications for M. bovis control at the wildlife–livestock interface
We provide supportive evidence of a multi-host transmission cycle of M. bovis in the Ruaha ecosystem, involving spillover between livestock and wildlife, as well as pathogen persistence in wildlife maintenance hosts. Unfortunately, eradication strategies in and around other protected areas where similar pathogen dynamics have been documented involve resource-intensive actions, such as fencing, culling, or large-scale test-and-slaughter programmes, which are not compatible with larger ecosystem conservation goals. Even with intense interventions, these efforts have met with limited success due to logistical challenges and the existence of other possible maintenance hosts that preclude a single-species intervention approach [Reference Renwick, White and Bengis33].
Perhaps what is most achievable in an ecosystem like Ruaha is to recognize that bTB is a multi-host disease affecting both wildlife and livestock and that a myriad of factors including spatio-temporal population overlap, animal density, drought, human-induced habitat change, and cultural practices will affect bTB transmission and should be considered when designing efforts to control it. An alternative approach to control and better understand bTB in this resource-poor system could include targeting testing of cattle and wildlife in shared grazing lands to identify areas or sites with increased spillover risk for management. For example, spatial analysis revealed two areas of higher than expected numbers of M. bovis infections within our sampling area. Even though these infection clusters were not statistically significant, potentially due to small sample size and low infection proportion, they may indicate a localized region within our sampling area that warrants additional field investigation. If resources were available to support more systematic bTB surveillance in both wildlife and livestock, focused testing in high-risk areas identified by spatial statistics could reveal a source of spillover or a highly infected local livestock or wildlife population for targeted intervention. Additionally, a better understanding of livestock grazing strategies, locations, and densities would also help identify transmission risk and control points.
At this time, there is no large-scale control or eradication programme for bTB in Tanzanian cattle or wildlife. Accordingly, the best approach to control the disease may be to focus on preserving the ecological functions of both protected and grazing areas to minimize both species overlap and the ecological stressors that increase an animal's susceptibility to bTB. For example, provision of adequate grazing and water resources for livestock would help minimize livestock incursions into wildlife protected areas during times of resource scarcity and would reduce inter-species transmission of not only bTB but potentially many other diseases. Programmes to facilitate market access for rural cattle could be coupled with efforts to promote sustainable livestock herd densities to optimize health, carcass condition, and efficient use of grazing resources. For wildlife population resilience, restoration of sustainable agricultural practices could support adequate dry-season water resources and minimize habitat degradation and encroachment.
CONCLUSION
A cattle strain of M. bovis infects multiple wildlife species inhabiting protected areas at the livestock–wildlife interface in the Ruaha ecosystem of Tanzania. Determining the species and spatial distribution of infection in both wildlife and livestock could enable an ecosystem-based approach to reduce disease transmission and improve opportunities for conservation interventions, tourism growth, livestock productivity, and livestock and human health security.
ACKNOWLEDGEMENTS
This research is part of the Health for Animals and Livelihood Improvement (HALI) Project, a One Health project addressing zoonotic diseases in Tanzania, and was made possible through support provided to the Global Livestock Collaborative Research Support Programme by the Office of Agriculture, Bureau for Economic Growth, Agriculture and Trade, United States Agency for International Development (USAID) under the terms of grant no. PCE-G-00-98-00036-00. The opinions expressed herein are those of the author(s) and do not necessarily reflect the views of the USAID. Additional funding for serological testing of buffalo was provided by the National Institute of Allergy and Infectious Diseases at the National Institutes of Health, International Collaborations in Infectious Disease Research grant no. U01AI088679.
We extend our gratitude to the game scouts, rangers and officers of the Matumizi Bora ya Malihai Idodi na Pawaga Association, the Iringa District Game Office, and Ruaha National Park for facilitating access to wildlife protected areas and samples; and to the South African Development Community and Tanzanian Ministry of Livestock for permission to test buffalo samples. We sincerely thank Ali Kitime, Jonas Fitwangile, Julis John, Joseph Malakalinga, and Lukiko Ndaki for laboratory assistance; Shukuru Mvena and Coaster Masimba for additional field assistance; Godwelias Ole Meing'ataki for field and logistical assistance within Ruaha National Park; Bakari Mbano and Ayubu Msago for their guidance and research facilitation; and Kate Thomas for help in creating the map in this paper. This paper is dedicated to the late Ali Kitime; who's friendship, laboratory mentorship, and countless teaching and research contributions are sorely missed.
DECLARATION OF INTEREST
None.





