Introduction
In all organisms, RNA polymerases (RNAP) are multimeric enzymes responsible for synthesizing RNA molecules. In eukaryotes, nuclear genomes are transcribed by DNA-dependent RNAP I, II and III (Roeder, Reference Roeder2019). RNAP I produces ribosomal RNAs (rRNAs) 18S, 5.8S and 28S, whereas RNAP II transcribes multiple types of RNA molecules, including messenger RNAs (mRNAs), most small nuclear RNAs (snRNAs) and small nucleolar RNAs (snoRNAs) (Liu et al., Reference Liu, Bushnell and Kornberg2013). RNAP III mediates the synthesis of structured, small RNAs such as transfer RNAs (tRNAs), 5S rRNA, U6 snRNA and 7SL RNA, many of which have functions related to protein synthesis and RNA processing (Dieci et al., Reference Dieci, Bosio, Fermi and Ferrari2013).
In yeast and vertebrates, most RNAP III promoters consist of sequence elements located downstream of the transcription start sites within the transcribed region, and they are classified into 3 main classes (Dieci et al., Reference Dieci, Fiorino, Castelnuovo, Teichmann and Pagano2007; Leśniewska and Boguta, Reference Leśniewska and Boguta2017). Type I promoters are typical of 5S rRNA genes, and they consist of 3 internal domains: Box A, an intermediate element and Box C. tRNA genes contain type II promoters that possess 2 conserved internal elements, Boxes A and B. Type III promoters, characteristic of U6 snRNA genes, consist of elements that reside exclusively upstream of the coding sequence: a TATA box, a proximal sequence element and a distal sequence element (Dieci et al., Reference Dieci, Bosio, Fermi and Ferrari2013).
General transcription factors TFIIIA, TFIIIB and TFIIIC are required by RNAP III to initiate RNA synthesis (Geiduschek and Kassavetis, Reference Geiduschek and Kassavetis2001). While TFIIIA is a single-subunit factor involved exclusively in the 5S rRNA production, TFIIIB participates in the transcription of all RNAP III-dependent genes; it is composed of the TATA-binding protein (TBP), the TFIIB-related factor 1 (Brf1) and B double prime 1 (Bdp1). TFIIIC is a 6-protein complex required for tRNA and 5S rRNA transcription. Another protein that regulates RNAP III activity is the transcriptional repressor Maf1 (Graczyk et al., Reference Graczyk, Cieśla and Boguta2018).
RNAP III is the largest of all eukaryotic RNAP, as it is composed of 17 different subunits (in comparison to 12 and 14 subunits for RNAP II and I, respectively) with a molecular weight of 0.7 MDa (Girbig et al., Reference Girbig, Misiaszek, Vorländer, Lafita, Grötsch, Baudin, Bateman and Müller2021). Five subunits are shared between all 3 RNAPs: RPB5, RPB6, RPB8, RPB10 and RPB12. Subunits AC40 and AC19 are common to RNAP I and III, and another 5 subunits (C160, C128, C25, C17 and C11) are homologous to RNAP I and II subunits. The remaining 5 subunits are specific to RNAP III, as they do not have structurally equivalent subunits in RNAP I or RNAP II. They form 2 stable subcomplexes, the C82/C34/C31 heterotrimer and the C53/C37 heterodimer (Geiduschek and Kassavetis, Reference Geiduschek and Kassavetis2001; Hu et al., Reference Hu, Wu, Sun, Yuan, Kobayashi, Myers and Hernandez2002).
The C82/C34/C31 heterotrimer is critical for specific transcription initiation at RNAP III-dependent genes, as it contributes to promoter opening (Wang and Roeder, Reference Wang and Roeder1997). Moreover, the C82/C34/C31 subcomplex seems to participate in the stabilization of the transcription bubble during RNA elongation (Lefèvre et al., Reference Lefèvre, Dumay-Odelot, El-Ayoubi, Budd, Legrand, Pinaud, Teichmann and Fribourg2011). C82, also known as RPC82 or RPC3 (and as RPC62 in humans), is characterized by the presence of 4 extended winged-helix (eWH) domains and a C-terminal coiled-coil domain. The eWH regions are required for the interactions that C82 establishes not only with C34 and C31, but also with C160 and C128, and with the Brf1 subunit of TFIIIB (Boissier et al., Reference Boissier, Dumay-Odelot, Teichmann and Fribourg2015; Hoffmann et al., Reference Hoffmann, Jakobi, Moreno-Morcillo, Glatt, Kosinski, Hagen, Sachse and Müller2015; Khoo et al., Reference Khoo, Wu, Lin and Chen2018). C82 also binds to promoter regions (Lefèvre et al., Reference Lefèvre, Dumay-Odelot, El-Ayoubi, Budd, Legrand, Pinaud, Teichmann and Fribourg2011; Khoo et al., Reference Khoo, Wu, Lin and Chen2018; Vorländer et al., Reference Vorländer, Khatter, Wetzel, Hagen and Müller2018), and displays helicase activity to unwind double-stranded DNA in an ATP-dependent fashion (Ayoubi et al., Reference Ayoubi, Dumay-Odelot, Chernev, Boissier, Minvielle-Sébastia, Urlaub, Fribourg and Teichmann2019). Thus, C82 plays key roles in RNAP III preinitiation complex formation and transcription initiation and elongation.
The parasitic protozoa Trypanosoma brucei and Leishmania spp. are members of the trypanosomatid family that affect millions of people worldwide. Trypanosoma brucei is endemic to Sub-Saharan Africa, where it produces sleeping sickness, also called human African trypanosomiasis (Pays et al., Reference Pays, Radwanska and Magez2023). The parasite is transmitted to humans by tsetse flies of the Glossina genus. Several species of Leishmania produce different types of leishmaniases in around 98 countries on 5 continents (Alvar et al., Reference Alvar, Vélez, Bern, Herrero, Desjeux, Cano, Jannin and den Boer2012; Singh et al., Reference Singh, Kashif, Srivastava and Manna2023). The pathogen is spread to humans by the bite of infected female sandflies of the genera Phlebotomus and Lutzomyia. Notably, trypanosomatids exhibit gene expression processes that are atypical among eukaryotes, such as RNAP II polycistronic transcription and mRNA maturation by trans-splicing (Martínez-Calvillo et al., Reference Martínez-Calvillo, Vizuet-de-Rueda, Florencio-Martínez, Manning-Cela and Figueroa-Angulo2010; Clayton, Reference Clayton2019).
In trypanosomatids, the knowledge about RNAP III transcription is scarce. Except for C31, orthologues of all RNAP III subunits have been identified in these parasites by tandem affinity purifications (Martínez-Calvillo et al., Reference Martínez-Calvillo, Saxena, Green, Leland and Myler2007; Florencio-Martínez et al., Reference Florencio-Martínez, Cano-Santiago, Mondragón-Rosas, Gómez-García, Flores-Pérez, Román-Carraro, Barocio-Rodríguez, Manning-Cela, Nepomuceno-Mejía and Martínez-Calvillo2021) or in silico analyses (El-Sayed et al., Reference El-Sayed, Myler, Blandin, Berriman, Crabtree, Aggarwal, Caler, Renauld, Worthey, Hertz-Fowler, Ghedin, Peacock, Bartholomeu, Haas, Tran, Wortman, Alsmark, Angiuoli, Anupama, Badger, Bringaud, Cadag, Carlton, Cerqueira, Creasy, Delcher, Djikeng, Embley, Hauser, Ivens, Kummerfeld, Pereira-Leal, Nilsson, Peterson, Salzberg, Shallom, Silva, Sundaram, Westenberger, White, Melville, Donelson, Andersson, Stuart and Hall2005; Kelly et al., Reference Kelly, Wickstead and Gull2005; Das et al., Reference Das, Banday and Bellofatto2008). Unlike other eukaryotes, trypanosomatid RNAP III transcribes not only the U6 snRNA gene, but the whole set of snRNA genes (Fantoni et al., Reference Fantoni, Dare and Tschudi1994). Interestingly, Boxes A and B located within neighbouring tRNA genes control the expression of snRNA genes in T. brucei and L. major (Nakaar et al., Reference Nakaar, Günzl, Ullu and Tschudi1997; Rojas-Sánchez et al., Reference Rojas-Sánchez, Figueroa-Angulo, Moreno-Campos, Florencio-Martínez, Manning-Cela and Martínez-Calvillo2016). Regarding general transcription factors, subunits TBP, Brf1 and Bdp1 from TFIIIB are essential proteins required for transcription initiation of RNAP III-dependent genes in L. major and T. brucei (Thomas et al., Reference Thomas, Yu, Sturm and Campbell2006; Vélez-Ramírez et al., Reference Vélez-Ramírez, Florencio-Martínez, Romero-Meza, Rojas-Sánchez, Moreno-Campos, Arroyo, Ortega-López, Manning-Cela and Martínez-Calvillo2015; Román-Carraro et al., Reference Román-Carraro, Florencio-Martínez, Romero-Meza, Nepomuceno-Mejía, Carrero, Arroyo, Ortega-López, Manning-Cela and Martínez-Calvillo2019; Florencio-Martínez et al., Reference Florencio-Martínez, Cano-Santiago, Mondragón-Rosas, Gómez-García, Flores-Pérez, Román-Carraro, Barocio-Rodríguez, Manning-Cela, Nepomuceno-Mejía and Martínez-Calvillo2021). Also, a 4-subunit TFIIIC complex that associates with tRNA and U2 snRNA genes was recently identified in trypanosomatids (Mondragón-Rosas et al., Reference Mondragón-Rosas, Florencio-Martínez, Villa-Delavequia, Manning-Cela, Carrero, Nepomuceno-Mejía and Martínez-Calvillo2024). The transcriptional repressor Maf1 (Romero-Meza et al., Reference Romero-Meza, Vélez-Ramírez, Florencio-Martínez, Román-Carraro, Manning-Cela, Hernández-Rivas and Martínez-Calvillo2017) and the SNAP50 subunit of the SNAP complex (Thomas et al., Reference Thomas, Green, Sturm, Campbell and Myler2009) have also been implicated in the regulation of RNAP III transcription in trypanosomatids.
In this work, we studied the C82 subunit of RNAP III in T. brucei (TbC82) and L. major (LmC82). While TbC82 and LmC82 possess the characteristic domains found in other C82 orthologues, they contain some unique features, including a trypanosomatid-specific loop within the eWH1 domain. Depletion of TbC82 by RNAi led to the growth arrest of the parasite, demonstrating that TbC82 is essential for T. brucei viability. Putative interacting partners of C82 were identified by tandem affinity purifications and mass spectrometry analyses, revealing similarities and differences between trypanosomatids and other organisms. Interestingly, some differences were also found between T. brucei and L. major.
Materials and methods
In silico analyses
Protein sequences were obtained from the TriTrypDB database (release 62) (http://tritrypdb.org/tritrypdb/) and the NCBI database (http://www.ncbi.nlm.nih.gov). Sequence alignments were made with the ClustalΩ program (http://www.ebi.ac.uk/Tools/msa/clustalo/) and shaded manually. Domain identification and secondary structure predictions were generated with the PSIPRED server (http://bioinf.cs.ucl.ac.uk/psipred/) and the Phyre2 program (http://www.sbg.bio.ic.ac.uk/~phyre2). The predicted 3-dimensional structures were generated with the AlphaFold program (https://alphafold.ebi.ac.uk/). Models were visualized and edited with the UCFS Chimera package (https://www.cgl.ucsf.edu/chimera/). Comparisons of the predicted structures were carried out with the iCn3D program (https://www.nlm.nih.gov/ncbi/workshops/2023-03_3d-molecular-structures/icn3d_alphafold.html). Hypothetical proteins identified by mass spectrometry were analysed with the HHpred program (https://toolkit.tuebingen.mpg.de/tools/hhpred), and the DALI server (http://ekhidna2.biocenter.helsinki.fi/dali/).
Cell culture and electroporation of L. major and T. brucei
Promastigotes from L. major strain MHOM/IL/81/Friedlin (LSB-132.1) were grown in BM medium supplemented with 10% fetal bovine serum (Life Technologies Corporation, Grand Island, NY, USA) at 28°C (Florencio-Martínez et al., Reference Florencio-Martínez, Cano-Santiago, Mondragón-Rosas, Gómez-García, Flores-Pérez, Román-Carraro, Barocio-Rodríguez, Manning-Cela, Nepomuceno-Mejía and Martínez-Calvillo2021). Transfection with the episomal pLmC82-PTP vector was performed by electroporation as previously described (Florencio-Martínez et al., Reference Florencio-Martínez, Cano-Santiago, Mondragón-Rosas, Gómez-García, Flores-Pérez, Román-Carraro, Barocio-Rodríguez, Manning-Cela, Nepomuceno-Mejía and Martínez-Calvillo2021). Clones were obtained by spreading transfected cells on plates containing 0.7% Seaplaque GTG agarose (FMC Bioproducts, Philadelphia, PA, USA) in BM medium with 50 μg mL−1 G418.
Procyclic forms of T. brucei strain 29-13 (Wirtz et al., Reference Wirtz, Leal, Ochatt and Cross1999) were cultured at 28°C in SDM-79 medium supplemented with 10% fetal bovine serum (Life Technologies Corporation), 50 μg mL−1 hygromycin B (Sigma-Aldrich, Darmstadt, Germany) and 15 μg mL−1 G418 (Sigma-Aldrich). Cells were transfected by electroporation, as previously described (Vélez-Ramírez et al., Reference Vélez-Ramírez, Florencio-Martínez, Romero-Meza, Rojas-Sánchez, Moreno-Campos, Arroyo, Ortega-López, Manning-Cela and Martínez-Calvillo2015). Populations were selected with phleomycin (2.5 μg mL−1) for RNAi or with blasticidin (10 μg mL−1) for PTP (Prot C-TEV-Prot A) tagging (Schimanski et al., Reference Schimanski, Nguyen and Günzl2005). Clones were obtained by serial dilution in 96-well plates. RNAi induction was carried out by adding doxycycline (2 μg mL−1) to the medium. The same clone was grown in the absence of doxycycline (non-induced control). For growth curves, parasites were counted daily and diluted to 2 × 106 cells mL−1, and cumulative cell density was plotted.
Generation of plasmids
To obtain the pC-TbC82-PTP plasmid for PTP-tagging, a 725-bp fragment from the C-terminal end of the TbC82 gene (Tb927.2.2990) was amplified with primers TbC82-C-PTP-ApaI-5’ (5’-GGGCCCTAGCGCATCAATGCCGCTT) and TbC82-C-PTP-NotI-3’-GC (5’-GCGGCCGCGCGTAAAAGTCCAAAACTAG). The DNA fragment was cloned into the genome-integration pC-PTP-BLA plasmid (Schimanski et al., Reference Schimanski, Nguyen and Günzl2005) with the ApaI and NotI restriction sites. The vector was linearized with the XcmI enzyme before transfection. To produce the pLmC82-PTP vector, the complete LmC82 gene (LmjF.27.2600), without the terminal codon, was amplified by PCR with oligonucleotides C82-AgeI-5’ (5’-ATACCGGTATATTTCTCCTCAGCAGGACTC) and C82-XbaI-3’ (5’-ATTCTAGAAAAAAAATCAACAATCAGCAGC). The PCR product was cloned into the episomal pB6-PTP plasmid (Moreno-Campos et al., Reference Moreno-Campos, Florencio-Martínez, Nepomuceno-Mejía, Rojas-Sánchez, Vélez-Ramírez, Padilla-Mejía, Figueroa-Angulo, Manning-Cela and Martínez-Calvillo2016) digested with XmaI and XbaI. To obtain plasmid p2T7-TbC82 for RNAi assays, a 419-bp fragment from the TbC82 gene was amplified with primers TbC82-RNAi-F (5’-AGGATCCAAGCTTGAACAACTCAGCCCTCTT) and TbC82-RNAi-R (5’-ACTCGAGATTGTCACACCCGTCTCTCC) and cloned into the p2T7-177 vector (Wickstead et al., Reference Wickstead, Ersfeld and Gull2002) digested with XhoI and BamHI. Prior to transfection the vector was linearized with NotI. To produce plasmid pCold-TbC82, the entire TbC82 gene was amplified with primers TbC82-BamHI-F (5’-GGATCCATGCCACGGCGTGCTGAG) and TbC82-XbaI-R (5’-TCTAGAGTAAAAGTCCAAAACTAGC) and cloned into the BamHI and XbaI restriction sites of the pCold1 expression vector (Takara Bio Inc., San Jose, CA, USA). All vectors were verified by sequencing.
Western blot analysis
Whole-cell protein extracts were made as described previously (Florencio-Martínez et al., Reference Florencio-Martínez, Cano-Santiago, Mondragón-Rosas, Gómez-García, Flores-Pérez, Román-Carraro, Barocio-Rodríguez, Manning-Cela, Nepomuceno-Mejía and Martínez-Calvillo2021). To perform Western blots, 20 μg of protein were fractionated by 10% SDS-PAGE and blotted onto PVDF (polyvinylidene difluoride membranes (Bio-Rad, Hercules, CA, USA). Next, the membranes were incubated with rabbit primary monoclonal anti-Prot C antibodies (Delta Biolabs, Boise, ID, USA) with a 1:3000 dilution, or polyclonal β-tubulin antibody (Thermo Scientific, Waltham, MA, USA) with a 1:1500 dilution; and then with a horseradish peroxidase (HRP)-conjugated secondary antibody with a 1:4000 dilution and developed with the Immobilon Western Chemiluminescent HRP substrate (Merck-Millipore, Darmstadt, Germany). The detection of TbC82 was achieved with a polyclonal anti-TbC82 antiserum (with a 1:500 dilution).
Indirect immunofluorescence
The subcellular localization of the TbC82-PTP and LmC82-PTP proteins was analysed by indirect immunofluorescence assays as previously described (Nepomuceno-Mejía et al., Reference Nepomuceno-Mejía, Florencio-Martínez and Martínez-Calvillo2018). To detect TbC82-PTP, T. brucei cells were fixed with 4% paraformaldehyde and incubated with rabbit anti-Prot C antibody (Delta Biolabs) followed by secondary anti-rabbit antibody conjugated with Alexa-Fluor 488 (Life Technologies Corporation). Likewise, L. major cells were fixed with 4% paraformaldehyde and incubated with a rabbit anti-Prot C antibody. Then, a mouse anti-LmNop56 antibody was used as a nucleolar marker. Parasites were then treated with a mixture of secondary anti-rabbit antibody conjugated with Alexa-Fluor 488 and anti-mouse antibody conjugated with Alexa Fluor 568 (Life Technologies Corporation). DNA was stained with DAPI (4′,6-diamidine-2′-phenylindole dihydrochloride). Images were obtained with a Zeiss AxioImager A2 microscope and analysed with the ZEN 2012 software (Blue 217 edition) (Zeiss, Oberkochen, Germany).
Northern blot analysis
The abundance of the TbC82 mRNA after RNAi was analysed by Northern blot experiments. For that purpose, total RNA was extracted with the TRI reagent (Sigma-Aldrich), and 20 μg were run in an agarose-formaldehyde denaturing gel and transferred to Hybond-N nylon membrane (GE HealthCare, Chicago, IL, USA). The radioactive probe employed corresponded to the 419-bp fragment cloned into the p2T7-TbC82 plasmid, labelled with [α-32P]-dCTP using the High Prime DNA Labelling Kit (Roche, Basel, Switzerland). Membranes were hybridized with a solution of formamide 50%, saline-sodium citrate (SSC) buffer 5×, sodium dodecyl sulphate (SDS) 0.2%, Denhardts 4× and salmon sperm DNA (100 μg mL−1) at 42°C, and then washed to a final stringency of 0.1 × SSC and 0.1% SDS at 65°C.
Tandem affinity purifications and mass spectrometry analysis
The proteins that associate with TbC82 and LmC82 were determined by performing tandem affinity purification assays, in duplicate, with parasites from the TbC82-PTP and LmC82-PTP cell lines (3 L at 2–3 × 107 cells mL−1) as previously described (Florencio-Martínez et al., Reference Florencio-Martínez, Cano-Santiago, Mondragón-Rosas, Gómez-García, Flores-Pérez, Román-Carraro, Barocio-Rodríguez, Manning-Cela, Nepomuceno-Mejía and Martínez-Calvillo2021). After the second column, the eluted proteins were concentrated with Amicon Ultra 3 K columns (Merck-Millipore) and by evaporation in a vacuum concentrator. Proteins were then analysed by SDS–PAGE and SYPRO Ruby (Invitrogen, Carlsbad, CA, USA) staining. Individual lanes from the gels were sliced into 2 pieces and proteins subjected to in-gel tryptic digestion prior to liquid chromatography–mass spectrometry/mass spectrometry at the Core Facility for Proteomics and Mass Spectrometry from Upstate Medical University (Syracuse, NY, USA). The collision-induced dissociation spectra were compared with the T. brucei and L. major protein database from the TriTrypDB page.
Chromatin immunoprecipitation (ChIP) assays
ChIP assays were carried out 3 times with the TbC82-PTP and LmC82-PTP cell lines, as described previously (Romero-Meza et al., Reference Romero-Meza, Vélez-Ramírez, Florencio-Martínez, Román-Carraro, Manning-Cela, Hernández-Rivas and Martínez-Calvillo2017). Briefly, 1.2 × 108 cells were cross-linked with 1% formaldehyde for 5 min at 37°C, and lysed with a Vibra-Cell VCX130 ultrasonic processor (Sonics, Newtown, CT, USA) (15 s on/off, 40% amplitude, for 3 min). Nuclei were pelleted and resuspended in sonication buffer (1% SDS, 10 mm EDTA and 50 mm Tris-HCl, pH 8.0, with 1× protease inhibitors). Chromatin was sonicated with a BioRuptor UCD-200 (Diagenode, Denville, NJ, USA) (30 s on/30 s off, high intensity) for 40 cycles, to an average DNA size of around 200–500 bp. The sonicated material was pre-cleared with protein A/G plus-agarose beads (Santa Cruz Biotechnology, Dallas, TX, USA) by mixing for 1 h at 4°C. Chromatin samples were incubated overnight at 4°C with rabbit anti-Prot A antibody (Sigma-Aldrich) or non-specific rabbit serum as negative control. The protein–DNA complexes were incubated for 2 h with protein A/G plus-agarose beads and 20 μg of sonicated salmon sperm DNA, and then washed as previously described (Vizuet-de-Rueda et al., Reference Vizuet-de-Rueda, Florencio-Martínez, Padilla-Mejía, Manning-Cela, Hernández-Rivas and Martínez-Calvillo2016). The cross-links were reversed with 200 mm NaCl at 65°C overnight and then treated with RNase A and proteinase K. DNA was precipitated with sodium acetate and ethanol and quantified.
Quantitative PCR assays
Quantitative PCR (qPCR) was performed with 2 ng of immunoprecipitated DNA to identify the regions of DNA to which TbC82 and LmC82 bind. The reactions were performed in duplicate, using optimized primers and conditions that produce a single amplicon of the correct size, with the Platinum SYBR Green qPCR SuperMix-UDG kit (Invitrogen). Results were analysed with the 2−ΔΔCq method, as reported before (Vélez-Ramírez et al., Reference Vélez-Ramírez, Florencio-Martínez, Romero-Meza, Rojas-Sánchez, Moreno-Campos, Arroyo, Ortega-López, Manning-Cela and Martínez-Calvillo2015; Vizuet-de-Rueda et al., Reference Vizuet-de-Rueda, Florencio-Martínez, Padilla-Mejía, Manning-Cela, Hernández-Rivas and Martínez-Calvillo2016), and are presented as percentage of input, corrected by subtracting the corresponding values from negative control precipitations performed with a non-specific antiserum. The upstream control region of the rRNA promoter (18S rRNA prom) of T. brucei was amplified with primers 18SUSE5 (5’-CACCCTCAAGACCGTAGCTC) and 18SUSE3 (5’-ACCCGTCCCTTATCAACACA). The 18S rRNA gene (Tb927.2.1452) was amplified with oligonucleotides 18SqFw (5’-GGGATACTCAAACCCATCCA) and 18SqRv (5’-CCCTTTAACAGCAACAGCATTA); and the α-tubulin gene (Tb927.1.2340) with TubqFw (5’-GGGCTTCCTCGTGTATCA) and TubqRv (5’-GCTTGGACTTCTTGCCATAG). The promoter of the SL gene (SL prom) was amplified with oligonucleotides SL-promoter-F (5’-CTACCGACACATTTCTGGC) and SL-promoter-R (5’-GCTGCTACTGGGAGCTTCTCATACC). The 5S rRNA gene (Tb927.8.1381) was amplified with primers rRNA5S-5′ (5’-GTCGAGTACGACCACACTTG) and rRNA5S-3’ (5’-AAGAGTACGGCACTCAGGGT). The tRNA-Ala gene (Tb927.7.6821) was amplified with primers AlaqFw (5’-GGGGATGTAGCTCAGATGG) and AlaqRv (5’-TGGAGAAGTTGGGTATCGATC); and the tRNA-Arg gene (Tb927.8.2859) with primers ArgqFw (5’-GGTCTCGTGGCGCAATG) and ArgqRv (5’-CGATCCCGGCAGGACTC). The intergenic region upstream of the tRNA-Ala gene (tRNA Ala inter) was amplified with primers InterAla5’ (5’-CACTCTCCCGAGAATCGAAG) and InterAla3’ (5’-TGGGTGTGGAGTCGACTTTT). The U2 snRNA gene (Tb927.2.5680) was amplified with primers U2qFw (5’-CTCGGCTATTTAGCTAAGATCAAGT) and U2qRv (5’-CGGGACAGCCAACAGTTT); and its promoter region (U2 snRNA prom) with primers U2Prom5’ (5’-CACAACCTGTAGTGGCGGTA) and Tb-U2-R (5’-GCATATCTTCTCGGCTATT).
For L. major, the 18S rRNA promoter (18S rRNA prom) was amplified with oligonucleotides rRNA-A-5’ (5’-TTGTTTGGGTGGAGGTGAGA) and rRNA-A-3’ (5’-CAAAATCATCAAACCCGTTC); and the rRNA 18S gene was amplified with primers rRNA-18S-5’ (5’-CATGCATGCCTCAGAATCAC) and rRNA-18S-3’ (5’-CGTTTCGCCAAGTTATCCAA). The protein-coding gene LmjF.11.0930 was amplified with primers 11.0930-5’ (5’-AGCAGCAGTTCATTGAGGCT) and 11.0930-3’ (5’-GCCGATCATCATCCTCTAAG); and the intergenic region upstream of LmjF.11.0930 (Inter 11.0930) was analysed with oligonucleotides 11-Inter-5’ (5’-GAACTTGGGAATGCCTTCTG) and 11-Inter-3’ (5’-GCAAGAAGAATGTGGAACGG). The amplification of SL promoter (SL prom) was carried out with primers LmjF-SL-PromF (5’-GAGCGCGGTGGGCATGACA) and LmjF-SL-PromR (5’-AAGCCATCACCACCGCAGC); and the intergenic region of the SL gene (Inter SL) was amplified with oligonucleotides LmjF-SL-InterF (5’-TGTGCGTGCGTGTGGTGGT) and LmjF-SL-InterR (5’-CGGGCGCACCCTTGCAGT). The strand-switch region (SSR) of chromosome 1 (SSR Chr. 1) was amplified with primers Ssr4-F (5’-AATCACAGCACGCATACACG) and Ssr4-R (5’-GCGTCATGGCTTCACTAACAG). The 5S rRNA gene was amplified with oligonucleotides 5SrRNA-F1 (5’-GAGTACGACCACACTTGAGTG) and 5SrRNA-R1 (5’-GAGTACGGCACTCAGGGTT). The U2 snRNA was analysed with primers U2-5’ (5’-AAACGTGGAACTCCAAGGAA) and U2-3’ (5’-TATCTTCTCGGCTATTTAGC); and the promoter region of the U2 snRNA (U2 snRNA prom) with primers U2tRNA-like-5’ (5’-CCGAGAAGATATGTTAGTACCACC) and U2tRNA-like-3’ (5’-AGGAAAAGATGCTTTCGACGAG). The tRNA-Met gene was amplified with oligonucleotides tRNAmet-F (5’-AAAGTTTGCGACCGGTGAG) and tRNAmet-R (5’-CACAACTTTCACTCGTAGCCG).
Reverse transcription qPCR
The abundance of different transcripts after TbC82 depletion by RNAi was analysed by reverse transcription qPCR (RT-qPCR) assays. Three biological replicates were examined. Briefly, 1 μg of total RNA from the induced and non-induced cultures was used as template for the first strand cDNA synthesis using the SuperScript III Reverse Transcriptase (Invitrogen) and 50 ng of random hexamers (Invitrogen). The cDNA was analysed by qPCR using the Platinum SYBR Green qPCR SuperMix-UDG kit (Invitrogen). The qPCR reactions were performed in duplicate. The procyclin transcript (Tb927.6.510) was amplified with primers Procyclin-5’ (5’-ATGGCACCTCGTTCCCTTTA) and ProcqRv (5’-CTTTGCCTCCCTTCACGATAAC); and TFIIB (Tb927.9.5710) with primers Tf2bqFw (5′-GAACAGGGAACGCACATTAG) and Tf2bqRv (5′-TTGTTGACTTTGGTCACTTCC). The 5S rRNA (Tb927.8.1381) was amplified with primers rRNA5S-5′ (5’-GTCGAGTACGACCACACTTG) and 5S rRNA-3’ (5’-TGAGCCTGTGAGTGCTTAACTT); and the TbC82 transcript was analysed with primers TbC82-GFP-F (5’-AGGTACCACCGGCTTCCAAAGAACT) and TbC82-C-PTP-NotI-3 (5’-GCGGCCGCGTAAAAGTCCAAAACTAGCATC). The α-tubulin, tRNA-Ala, tRNA-Arg and U2 snRNA were amplified with the primers mentioned in the previous section.
Production of TbC82 polyclonal antibody
Escherichia coli BL21 (DE3) competent cells (Thermo Scientific) were transformed with the pCold-TbC82 plasmid. Induction of the TbC82 recombinant protein (TbC82r) expression was achieved with 1 mm isopropyl β-D-1-thiogalactopyranoside at 37°C for 18 h. The TbC82r protein was purified by affinity chromatography with Ni-Sepharose 6 Fast Flow matrix (GE Healthcare), according to the manufacturer's instructions. The anti-TbC82 polyclonal antibody was produced by inoculating 6-week-old male BALB/c mice intravenously with 100 μg of purified TbC82r protein mixed with TiterMax Gold adjuvant (Sigma-Aldrich) at a 1:1 ratio. Pre-immune mouse serum was obtained before antigen inoculation. Blood samples were collected 6 weeks after antigen immunization, and anti-TbC82 polyclonal serum was recovered by centrifugation. The specificity of the anti-TbC82 antibody was confirmed by Western blot analysis.
Results
C82 exhibits some distinctive features in trypanosomatids
The proteins encoded by Tb927.2.2990 and LmjF.27.2600 were identified as the C82 subunits of RNAP III in T. brucei (TbC82) and L. major (LmC82), respectively (Martínez-Calvillo et al., Reference Martínez-Calvillo, Saxena, Green, Leland and Myler2007). Sequence comparisons show that human C82 (RPC62) is 13.64% identical to TbC82 and 16.05% identical to LmC82 (Figs 1A and S1). This low sequence identity is not surprising, since the conservation of C82 orthologues is low across eukaryotes, with 19.25% identity between humans and Saccharomyces cerevisiae, and 25.78% identity between Schizosaccharomyces pombe and S. cerevisiae (Martínez-Calvillo et al., Reference Martínez-Calvillo, Saxena, Green, Leland and Myler2007) (Fig. S1). Regardless of the low sequence homology, both TbC82 and LmC82 contain the 4 characteristic eWH domains, and the coiled-coil domain in the C-terminal region (Fig. 1). While typical WH folds are composed of 3 α-helices and 3 β-strands in the order α1-β1-α2-α3-β2-β3, eWH domains in C82 possess an extra α-helix (α0) at the N terminus, the best fit being found for the eWH1 domain (Lefèvre et al., Reference Lefèvre, Dumay-Odelot, El-Ayoubi, Budd, Legrand, Pinaud, Teichmann and Fribourg2011) (Fig. 1A). However, a distinctive characteristic is that the insertion loop, which interrupts eWH2, is shorter in trypanosomatids (Fig. 1A). Moreover, these organisms contain an insertion within the eWH1 domain, which is not present in the other species analysed, and that we named ‘trypanosomatid-specific loop’ (Figs 1A and S1B).

Figure 1. Sequence and predicted 3-dimensional structure analyses of C82 in T. brucei and L. major. (A) Alignment of the complete C82 amino acid sequences of T. brucei (Tb, Tb927.2.2990), L. major (Lm, LmjF27.2600) and H. sapiens (Hs, RPC62, NP_006459.3). The location of the 4 extended winged-helix (eWH) domains and the coiled-coil (C. coil) motif are indicated. Conserved residues are denoted by black shading, conserved substitutions by dark-grey shading and semiconserved substitutions by light-grey shading, according to the Clustal Ω program. The predicted secondary structure elements are shown for T. brucei (above the sequence) and H. sapiens (below the sequence). The α-helices are indicated with rectangles and the β-strands with arrows. (B) Predicted 3-dimensional structures of the entire TbC82 and LmC82 proteins generated with the AlphaFold program. For comparison, the structure of H. sapiens C82 (RPC62) is also presented. The structures are displayed in the same colours shown in panel A. Comparisons of the predicted structures with the iCn3D program revealed the following TM-scores: Tb/Lm = 0.740; Tb/Hs = 0.735; and Lm/Hs = 0.747.
As expected, C82 sequence conservation is higher among trypanosomatids, with identities ranging from 81.6 to 97.6% between Leishmania species, and from 57.8 to 80.2% between Trypanosoma species (Fig. S2). A 29.9% identity is observed between LmC82 and TbC82. To further analyse TbC82 and LmC82, their predicted 3-dimensional structure was generated with the AlphaFold program and compared to the structure of human C82. As shown in Fig. 1B, in all cases C82 is a globular protein mainly composed of α-helices, with 2 central long α-helices (the coiled-coil region) surrounded by the 4 well-defined eWH domains. Two clear differences are the presence of the longer insertion loop in the human eWH2, and the trypanosomatid-specific loop in the eWH1 of T. brucei and L. major. Notably, the predicted structure of TbC82 shows some differences when compared to the other 2 proteins and, consequently, the hypothetical structure of LmC82 is more similar to the human C82 structure than to the TbC82 structure (Fig. 1B).
LmC82 and TbC82 localize to the nucleus
The subcellular localization of C82 was determined in L. major promastigotes and T. brucei procyclic forms by indirect immunofluorescence experiments. For that purpose, we produced cell lines where C82 was tagged with a carboxy-terminal PTP domain, which is composed of Protein A (Prot A) and Protein C (Prot C) epitopes separated by a TEV protease cleavage site (Schimanski et al., Reference Schimanski, Nguyen and Günzl2005). For L. major analysis, transgenic promastigotes that express the LmC82-PTP recombinant protein were produced with the pLmC82-PTP vector, obtained by cloning the complete LmC82 coding region into the episomal plasmid pB6-PTP (Moreno-Campos et al., Reference Moreno-Campos, Florencio-Martínez, Nepomuceno-Mejía, Rojas-Sánchez, Vélez-Ramírez, Padilla-Mejía, Figueroa-Angulo, Manning-Cela and Martínez-Calvillo2016).
The expression of the LmC82-PTP protein was verified by Western blot experiments carried out with an anti-Prot C antibody, showing the expected band of ~83 kDa, product of the fusion of the 20 kDa PTP tag with the ~63 kDa LmC82 (Fig. 2A). Indirect immunofluorescence assays carried out with the anti-Prot C antibody showed that LmC82 is localized to the nucleus of the transfected parasites (Fig. 2B). While most signal was observed in the nucleoplasm, some fluorescence was found in the outer part of the nucleolus (Fig. 2B). In yeast, where 5S rRNA and tRNA genes are located within the nucleolus (Thompson et al., Reference Thompson, Haeusler, Good and Engelke2003), C82 is distributed throughout the entire nucleus (Wei et al., Reference Wei, Tsang and Zheng2009). For T. brucei, the 3’-terminal coding sequence of TbC82 was cloned into the genome-integration vector pC-PTP-BLA (Schimanski et al., Reference Schimanski, Nguyen and Günzl2005) to obtain the plasmid pC-TbC82-PTP. Western blot experiments with the anti-Prot C antibody allowed the identification of the expected ~81 kDa band (the predicted mass of TbC82 is ~61 kDa) (Fig. 3A). Like in L. major, indirect immunofluorescence experiments showed that TbC82-PTP is a nuclear protein in T. brucei (Fig. 3B). This is in agreement with a genome-wide study that mapped the subcellular fate of most T. brucei proteins, where they found a nuclear localization for C-terminally mNeonGreen-tagged TbC82 (Billington et al., Reference Billington, Halliday, Madden, Dyer, Barker, Moreira-Leite, Carrington, Vaughan, Hertz-Fowler, Dean, Sunter, Wheeler and Gull2023) (data available on the TrypTag website, http://tryptag.org). Collectively, these results show that the PTP tag did not affect the nuclear localization of C82 in L. major and T. brucei.

Figure 2. LmC82 is a nuclear protein. (A) Western blot analysis with parasites that express the LmC82-PTP protein and wild-type (WT) cells. Membranes were incubated with an antibody against Prot C and an anti-β-tubulin antibody (loading control). (B) Indirect immunofluorescence experiments to determine the subcellular localization of LmC82-PTP using an anti-Prot C antibody. An anti-LmNop56 antibody was used as a nucleolar marker. Parasites were then treated with a mixture of secondary anti-rabbit antibody conjugated with Alexa-Fluor 488 and anti-mouse antibody conjugated with Alexa Fluor 568 (Life Technologies Corporation). Nucleus (N) and kinetoplast (K) were stained with DAPI. Size bars represent 5 μm.

Figure 3. Subcellular localization of TbC82. (A) Western blot analysis with total protein from wild-type (WT) cells and parasites that express the TbC82-PTP protein using an anti-Prot C monoclonal antibody. As a loading control, β-tubulin was used. (B) The subcellular location of TbC82-PTP was analysed by indirect immunofluorescence assays using an anti-Prot C monoclonal antibody and an Alexa-Fluor 488 conjugated secondary antibody (Life Technologies Corporation). Nucleus (N) and kinetoplast (K) were stained with DAPI. Size bars represent 5 μm.
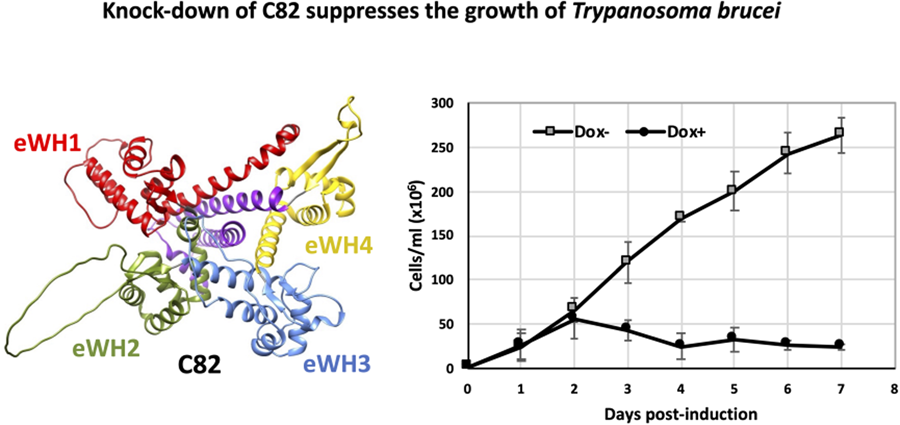
C82 is an essential protein in procyclic forms of T. brucei
To assess whether TbC82 is indispensable for T. brucei survival, TbC82 was knocked down in vivo by RNAi. To that end, plasmid p2T7-C82 was generated by cloning a 419-bp fragment from the TbC82 coding region into p2T7-177, a vector that contains 2 opposite tetracycline-inducible T7 RNAP promoters to produce double-stranded RNA (Wickstead et al., Reference Wickstead, Ersfeld and Gull2002). Plasmid p2T7-C82 was transfected into the procyclic T. brucei cell line 29–13, which expresses the tetracycline repressor and T7 RNAP (Wirtz et al., Reference Wirtz, Leal, Ochatt and Cross1999). The transfected population was cloned by limiting dilution, and a clonal cell line was selected for further analysis. To evaluate the effect of the TbC82 knock-down on the growth of T. brucei, cultures induced by the addition of the tetracycline analogue doxycycline (Dox+), and non-induced cultures (Dox–), were counted and diluted daily for 7 days. As shown in Fig. 4A, while Dox– parasites grew normally, Dox+ cells stopped growing 2 days after RNAi induction, leading to cell death 2 days later. To corroborate the TbC82 mRNA depletion after doxycycline induction, Northern blot analysis was performed, showing that the levels of the TbC82 mRNA were decreased by around 79% on day 2 post-induction (Fig. 4B). RT-qPCR experiments revealed that the levels of the TbC82 transcript were reduced by ~55% on day 4 post-induction (Fig. 5). Western blot analysis performed with a TbC82 polyclonal antiserum showed that the abundance of the TbC82 protein was reduced by approximately 48.5% after 3 days of induction (Fig. 4C). Thus, these results demonstrate that TbC82 is essential for the survival of procyclic forms of T. brucei.

Figure 4. C82 is essential for cell growth of procyclic forms of T. brucei. (A) Growth curve of a clonal cell line obtained with the p2T7-TbC82 vector under non-induced (Dox−) and doxycycline-induced (Dox+) conditions. Cells were counted daily and diluted to a density of 2 × 106 cells mL−1. The values represent the cumulative cell density multiplied by the dilution factor. Data points reflect the means of triplicate experiments. Standard deviation bars are shown. (B) Northern blot analysis of TbC82 mRNA in non-induced cells (0 days), and cells induced for 1, 2 or 3 days. The bands shown here and from 2 independent experiments were quantified and plotted, considering as 100% the RNA level obtained in the non-induced culture. Values represent means of the 3 experiments. Levels of TbC82 mRNA were normalized to the level of the α-tubulin mRNA (loading control). (C) Western blot analysis of the TbC82 protein in non-induced cells (0 days), and cells induced for 3 days using a specific anti-TbC82 polyclonal antibody. The bands shown here and from 2 independent experiments were quantified and plotted, considering as 100% the protein level obtained in the non-induced culture. Values represent means of the 3 experiments. Standard deviation bars are shown. TbC82 protein levels were normalized to the level of the β-tubulin protein (loading control).

Figure 5. Depletion of TbC82 decreases the abundance of RNAP III-dependent transcripts. Quantitative PCR analysis of total RNA from induced (for 3 and 4 days) and non-induced (Dox−) TbC82 RNAi cultures. The RNAP III-dependent transcripts analysed were tRNA Arg, tRNA Ala, 5S rRNA and U2 snRNA. As controls, we analysed Procyclin (transcribed by RNAP I) and TFIIB (transcribed by RNAP II). We also evaluated the TbC82 transcript. Three biological replicates were analysed. All qPCR reactions were performed in duplicate, using primers and conditions that were optimized to produce a single amplicon of the correct size. Error bars indicate standard deviations. Statistically significant differences (Tukey's test) compared to the Dox-culture are indicated with * (P < 0.05) or ** (P < 0.01).
Knock-down of TbC82 led to a reduction in the levels of RNAP III-dependent transcripts
The effect of the ablation of TbC82 on the abundance of RNA molecules synthesized by RNAP III was analysed by RT-qPCR. These experiments were performed with total RNA isolated from cultures induced for 3 and 4 days. As shown in Fig. 5, a strong reduction in the abundance of all RNAP III-dependent transcripts analysed was observed, especially on day 3 post-induction. While the abundance of the tRNA-Arg and tRNA-Ala decreased to 23 and 21% of the control values, respectively, 5S rRNA was reduced to 18%. The level of the U2 snRNA was also reduced, but to a lesser extent (44% after 3 days of induction). Thus, these results demonstrate the participation of TbC82 in the transcription of all types of RNAP III-dependent genes in T. brucei. The abundances of the mRNAs encoding procyclin (synthesized by RNAP I) and TFIIB (transcribed by RNAP II) were not affected by TbC82 knock-down (Fig. 5).
TbC82 and LmC82 associate with RNAP III-dependent genes
To demonstrate the in vivo binding of subunit C82 to genes transcribed by RNAP III in T. brucei and L. major, ChIP experiments were carried out with the clonal cell lines that express C82 fused to the PTP tag. Immunoprecipitations were performed with a ChIP-grade anti-Prot A antibody that recognizes the Prot A epitopes from the PTP tag, and with a non-specific mouse serum as a negative control. The association of TbC82-PTP and LmC82-PTP to the T. brucei and L. major genomes, respectively, was evaluated by qPCR assays conducted with the purified DNA. In T. brucei, strong occupancy of TbC82 was observed in the 5S rRNA, the tRNA-Arg and the tRNA-Ala genes (Fig. 6). The association of TbC82 was not found in an intergenic region located downstream of the tRNA-Ala gene. Enrichment of TbC82 was also detected in the U2 snRNA gene and its upstream promoter region. As anticipated, binding of TbC82 was not observed in the SL RNA promoter and the α-tubulin gene (transcribed by RNAP II) or in the 18S rRNA gene and its promoter region (transcribed by RNAP I) (Fig. 6). With L. major we obtained similar results, as high occupancy of LmC82 was found in the 5S rRNA and the tRNA-Met genes, as well as in the U2 snRNA gene and its upstream promoter region (Fig. 7). Binding of LmC82 was also observed in the intergenic region downstream of the 5S rRNA gene, but not in the neighbouring protein-coding gene LmjF.11.0930. Occupancy of LmC82 was not detected in the SSR from chromosome 1 and the SL RNA locus (transcribed by RNAP II), and the 18S rRNA gene and promoter region (transcribed by RNAP I) (Fig. 7). Altogether, these results show that C82 associates in vivo with RNAP III-dependent genes in T. brucei and L. major.

Figure 6. Chromatin immunoprecipitation analysis of TbC82. (A) Schematic drawing of the studied genes and amplicons quantified in panel B. Genomic regions transcribed by RNAP III, RNAP II and RNAP I are shown in blue, yellow and green, respectively. Maps are not shown to scale. (B) A ChIP grade anti-Prot A antibody was used to precipitate chromatin from the cell line that expresses the TbC82-PTP protein. Precipitated DNA was examined by qPCR. The results from 3 independent ChIP experiments, each analysing 2 qPCR reactions, are shown. Error bars indicate standard deviations. Results are presented as percentage of input, corrected by subtracting corresponding values from negative control precipitations performed with a nonspecific antiserum.

Figure 7. ChIP analysis of LmC82. (A) Genomic maps of the analysed loci. Regions transcribed by RNAP III, RNAP II and RNAP I are shown in blue, yellow and green, respectively. Maps are not shown to scale. (B) ChIP analysis using an anti-Prot A antibody was carried out with the cell line that expresses the LmC82-PTP protein. Precipitated DNA was examined by qPCR. The results from 3 independent ChIP experiments, each analysing 2 qPCR reactions, are shown. Error bars indicate standard deviations. Results are presented as percentage of input, corrected by subtracting corresponding values from negative control precipitations performed with a non-specific antiserum.
Proteins that participate in RNAP III transcription and other functions copurified with TbC82-PTP and LmC82-PTP
In order to determine the proteins that interact directly or indirectly with the C82 subunit in T. brucei and L. major, tandem affinity purification experiments were carried out with the cell lines that express the recombinant proteins TbC82-PTP and LmC82-PTP, respectively. Associated proteins were isolated by IgG affinity chromatography, TEV protease elution and anti-Prot C affinity chromatography. SDS-PAGE analyses of the eluted material showed multiple protein bands, including 2 of ~64 and ~66 kDa that seem to correspond to the Prot C-tagged versions of TbC82 (Fig. 8A) and LmC82 (Fig. 8B), respectively (denoted with asterisks). As controls, purifications using wild-type T. brucei and L. major extracts were performed. Electrophoretic analysis of these controls showed the presence of a low number of faint bands (Fig. 8, WT lanes) that were identified by mass spectrometry as bovine serum albumin, human keratins, and several trypanosomatid ribosomal proteins, heat shock proteins, translation elongation factors, mitochondrial proteins, α- and β-tubulins and some other proteins (Table S1). These proteins are common contaminants in tandem affinity purifications (Mellacheruvu et al., Reference Mellacheruvu, Wright, Couzens, Lambert, St-Denis, Li, Miteva, Hauri, Sardiu, Low, Halim, Bagshaw, Hubner, Al-Hakim, Bouchard, Faubert, Fermin, Dunham, Goudreault, Lin, Badillo, Pawson, Durocher, Coulombe, Aebersold, Superti-Furga, Colinge, Heck, Choi, Gstaiger, Mohammed, Cristea, Bennett, Washburn, Raught, Ewing, Gingras and Nesvizhskii2013).

Figure 8. Tandem affinity purifications with T. brucei and L. major parasites expressing C82-PTP recombinant proteins. SDS-PAGE of proteins copurified with TbC82-PTP (A) and LmC82-PTP (B). The asterisks indicate the PTP-fused proteins. As controls, experiments with wild-type (WT) T. brucei (A) and L. major (B) parasites were also conducted. Proteins were analysed in 4–15% Mini- PROTEAN Precast Protein Gels (Bio-Rad) stained with SYPRO Ruby (Invitrogen).
The TbC82-PTP and LmC82-PTP samples were examined by mass spectrometry and bioinformatic analyses, leading to the identification of numerous proteins (Table S1). To discriminate between contaminants and genuine C82 interactors, we applied the following criteria: the proteins identified on the wild-type cell purifications were considered contaminants, together with other proteins that we have usually detected in unrelated tandem affinity purifications (Florencio-Martínez et al., Reference Florencio-Martínez, Cano-Santiago, Mondragón-Rosas, Gómez-García, Flores-Pérez, Román-Carraro, Barocio-Rodríguez, Manning-Cela, Nepomuceno-Mejía and Martínez-Calvillo2021; Mondragón-Rosas et al., Reference Mondragón-Rosas, Florencio-Martínez, Villa-Delavequia, Manning-Cela, Carrero, Nepomuceno-Mejía and Martínez-Calvillo2024); also, proteins that presented an average coverage of less than 7.5% in T. brucei, or less than 20% in L. major, were regarded as possible contaminants. A lower coverage threshold was set for T. brucei because in this parasite we identified a smaller number of proteins, with lower overall protein coverages than those observed in L. major (Table S1). Accordingly, Tables 1 and 2 present the putative interacting partners of TbC82 and LmC82, respectively, which were grouped into 6 different categories: RNAP subunits, transcription factor TFIIIC subunits, putative transcription regulators, RNA-binding proteins, transport proteins and other functions. These proteins might associate directly or indirectly, via other proteins, with the C82 subunit. Since some DNA and RNA molecules could copurify with the tagged protein in affinity purifications, they may contribute to the indirect association of proteins with C82, and potentially influence the observed differences between T. brucei and L. major (see below).
Table 1. Proteins that copurified with TbC82a

a Proteins likely to be contaminants (including multiple ribosomal proteins, translation factors, tubulins, heat-shock proteins, mitochondrial proteins) were not included.
b Each digit indicates the number of peptides identified in 2 different tandem affinity purifications.
c Each number denotes the coverage found in 2 different experiments. Only proteins that show a coverage of at least 7.5%, in average, are shown.
Table 2. Putative interacting partners of LmC82a

a Proteins likely to be contaminants (including multiple ribosomal proteins, translation factors, tubulins, heat-shock proteins, mitochondrial proteins) were not included.
b Each digit indicates the number of peptides identified in 2 different tandem affinity purifications.
c Each number denotes the coverage found in 2 different experiments. Only proteins that show a coverage of at least 20%, in average, are shown.
d Around half of the identified peptides are shared with LmjF.020680, the smaller C82 isoform in L. major.
Regarding RNAPs, 4 RNAP III subunits (3 exclusive and 1 shared with RNAP I) copurified with TbC82, while 9 RNAP III subunits (either exclusive or shared with RNAP I or II), as well as an RNAP I subunit, were identified with LmC82. With TbC82, we also identified 4 TFIIIC subunits and 2 possible regulators of transcription, one of which is Trypanosoma-specific (Table 1). LmC82-PTP was copurified with a protein related to the TBP-Interacting Protein 120 (TIP120), the NLI interacting factor-like phosphatase, and a Leishmania-specific putative transcriptional regulator, together with several RNA-binding proteins and transport proteins (Table 2).
Discussion
In this work, we studied the C82 subunit of RNAP III in T. brucei and L. major. To the best of our knowledge, this work represents the first characterization of C82 in any protozoan species. Although TbC82 and LmC82 show secondary and 3-dimensional structure conservation (Fig. 1), they (as well as C82 orthologues in other trypanosomatids) contain a trypanosomatid-specific loop within the eWH1 domain, whose size ranges from 30 amino acids (in T. cruzi and T. rangeli) to 49 amino acids in Leishmania spp. (Figs S1B and S2A). It is also worth noting that the insertion loop located in the eWH2 domain is shorter in trypanosomatids (only 12 amino acids in Leptomonas seymouri and T. rangeli) (Fig. S2A) than in other eukaryotes (64 amino acids in human cells) (Fig. 1). Thus, it would be interesting to determine the implications that these differences may have on the function of C82 and the complete RNAP III complex in trypanosomatids.
Interestingly, L. major possesses a truncated copy of C82 (LmjF.02.0680) (hereafter referred to as LmC82-short), not present in Trypanosoma spp., which seems to have arisen as the result of a recombination event between the ends of chromosomes 2 and 27, and that was considered a pseudogene (Martínez-Calvillo et al., Reference Martínez-Calvillo, Saxena, Green, Leland and Myler2007). A sequence comparison between LmC82 (604 amino acids) and LmC82-short (501 amino acids) showed that the latter lacks the eWH1 domain but contains a C-terminal extension (Fig. S3A). Also, LmC82-short presents a highly divergent eWH2 motif, but very conserved eWH3, eWH4 and coiled-coil domains (Fig. S3A). Mass spectrometry analyses of the LmC82-PTP complexes (see below) revealed that 47.8% of the identified peptides are exclusive to LmC82, and 52.1% are shared between both C82 isoforms. Interestingly, we identified a peptide exclusive to LmC82-short (Fig. S3A), indicating that this gene is expressed at the protein level. Since C82 has not been reported to form homodimers, a direct association between LmC82 and LmC82-short is unlikely. Thus, it is possible that LmC82-short interacts with other RNAP III subunits and that it was indirectly copurified in our experiments. Future studies will help to determine if LmC82-short is required for RNAP III transcription in L. major. Of note, a truncated C82 gene is found in the majority of the Leishmania species whose genome sequences are deposited on the TriTrypDB website. In all these species, the predicted protein sequences are even smaller than LmC82-short (for instance, 158 amino acids in L. mexicana and 192 amino acids in L. donovani) (Figs S3B and S3C), indicating that they could actually represent pseudogenes.
In yeast, the C82 subunit of RNAP III is essential for cell viability (Chiannilkulchai et al., Reference Chiannilkulchai, Stalder, Riva, Carles, Werner and Sentenac1992). Accordingly, ablation of C82 by RNAi demonstrated that this subunit is indispensable for the survival of procyclic forms of T. brucei (Fig. 4). It should be noted that previous genome-wide RNAi knock-down screens reported that TbC82 is not essential in the procyclic and bloodstream stages of the parasite (Alsford et al., Reference Alsford, Turner, Obado, Sanchez-Flores, Glover, Berriman, Hertz-Fowler and Horn2011).
PTP-tag purifications with the TbC82 and LmC82 transgenic lines, followed by mass spectrometry and in silico analyses, led to the identification of multiple putative C82 interacting partners (Tables 1 and 2). While several RNAP III subunits were identified with both parasites, a RNAP I-specific subunit (RPB10z) was also copurified with LmC82. Among the RNAP III subunits, in both parasites we found C34, which in other species forms part of the RNAP III-specific heterotrimer (C82/C34/C31). In human cells, all 4 eWH domains from C82 (RPC62) seem to be required for the association with the C34 orthologue (RPC39) (Lefèvre et al., Reference Lefèvre, Dumay-Odelot, El-Ayoubi, Budd, Legrand, Pinaud, Teichmann and Fribourg2011; Li et al., Reference Li, Yu, Zhao, Ren, Hou and Xu2021). It is worth mentioning that C82 and C34 are structurally related to subunits TFIIEα and TFIIEβ, required for RNAP II transcription, and to archaeal TFEα/β, as they all contain multiple eWH motifs (Blombach et al., Reference Blombach, Salvadori, Fouqueau, Yan, Reimann, Sheppard, Smollett, Albers, Kay, Thalassinos and Werner2015; Khoo et al., Reference Khoo, Wu, Lin and Chen2018). Interestingly, we did not find an orthologue of the C31 subunit, the third component of the heterotrimer, and the only RNAP III subunit that has not been identified in trypanosomatids. In yeast and human cells, C31 possesses several conserved regions, including an Asp-Glu-rich acidic C-terminus, a pre-acidic domain, a conserved block and a stalk bridge helix (Boissier et al., Reference Boissier, Dumay-Odelot, Teichmann and Fribourg2015; Shekhar et al., Reference Shekhar, Sun, Khoo, Lin, Malau, Chang and Chen2023). None of the proteins that copurified with TbC82 and LmC82 seem to contain these domains, which suggests that C31 is absent in trypanosomatid parasites.
Remarkably, our results show that the degree of association between the TFIIIC subunits and C82 differs considerably between T. brucei and L. major. In the former, all 4 TFIIIC subunits that have been described in trypanosomatids (Mondragón-Rosas et al., Reference Mondragón-Rosas, Florencio-Martínez, Villa-Delavequia, Manning-Cela, Carrero, Nepomuceno-Mejía and Martínez-Calvillo2024) copurified with TbC82 with a high number of peptides and coverage (Table 1). For instance, with the Tau95 subunit, both the number of identified peptides (32 and 31 in experiments 1 and 2, respectively) and the average coverage (46.5%) were even greater than those found with the C34 subunit of RNAP III (14 and 13 peptides; 35% average coverage). A similar result was observed with the Tau55 subunit (22 and 20 peptides; 65.5% coverage). In contrast, LmC82 was copurified with only 3 TFIIIC subunits, and they showed a low number of peptides and coverage (consequently, they are not shown in Table 2). The identified subunits are Tau131 (3 and 7 peptides; 5.5% coverage), Tau138 (3 and 3 peptides; 2.5% coverage) and Tau95 (1 and 4 peptides; 4.5% coverage). Similar to T. brucei, for C34 in L. major we identified 15 and 19 peptides with an average coverage of 37.5% (Table 2). In support of these findings, tandem affinity purifications performed with cells expressing the Tau95-PTP recombinant protein showed a robust association between Tau95 and C82 in T. brucei, but not in L. major (Mondragón-Rosas et al., Reference Mondragón-Rosas, Florencio-Martínez, Villa-Delavequia, Manning-Cela, Carrero, Nepomuceno-Mejía and Martínez-Calvillo2024). Thus, altogether these results indicate that the interaction between the C82 subunit and transcription factor TFIIIC is strong in T. brucei, but weak in L. major. In human cells, a weak in vitro association between C82 (RPC62) and Tau95 (TFIIIC63) has been reported (Hsieh et al., Reference Hsieh, Wang, Kovelman and Roeder1999).
The robust copurification of all TFIIIC subunits with TbC82 is unexpected, taking into consideration that when RNAP subunits are used as baits in affinity purifications, the most abundant copurifying proteins are always other RNAP subunits (and not transcription factors), as reported in trypanosomatids (Walgraffe et al., Reference Walgraffe, Devaux, Lecordier, Dierick, Dieu, Van den Abbeele, Pays and Vanhamme2005; Das et al., Reference Das, Li, Liu and Bellofatto2006; Devaux et al., Reference Devaux, Lecordier, Uzureau, Walgraffe, Dierick, Poelvoorde, Pays and Vanhamme2006; Nguyen et al., Reference Nguyen, Schimanski, Zahn, Klumpp and Günzl2006; Martínez-Calvillo et al., Reference Martínez-Calvillo, Saxena, Green, Leland and Myler2007) and other organisms (Jeronimo et al., Reference Jeronimo, Langelier, Zeghouf, Cojocaru, Bergeron, Baali, Forget, Mnaimneh, Davierwala, Pootoolal, Chandy, Canadien, Beattie, Richards, Workman, Hughes, Greenblatt and Coulombe2004; Nguyen et al., Reference Nguyen, Saguez, Conesa, Lefebvre and Acker2015; Bhalla et al., Reference Bhalla, Vernekar, Gilquin, Couté and Bhargava2019), and as found here for LmC82 (Table 2). As an example, tandem chromatin affinity purifications with yeast cells expressing a tagged C160 subunit resulted in the copious copurification of most RNAP III subunits, but TFIIIC was not identified (Nguyen et al., Reference Nguyen, Saguez, Conesa, Lefebvre and Acker2015). Thus, the atypical strong association between C82 and TFIIIC in T. brucei requires further analysis.
Other proteins that copurified with TbC82 are Tb927.11.6310 and Tb927.11.10550 (Table 1), which are annotated as hypothetical proteins. The former is a nuclear trypanosome-specific protein (Billington et al., Reference Billington, Halliday, Madden, Dyer, Barker, Moreira-Leite, Carrington, Vaughan, Hertz-Fowler, Dean, Sunter, Wheeler and Gull2023) that, according to the HHpred server, presents weak homology to some proteins involved in transcription regulation, such as mitochondrial transcription termination factor 2 (mTERF-2) (probability of 69%, E-value of 12) and elongation factor SPT5 (probability of 39%, E-value of 37). The DALI server identified Tb927.11.6310 as a feasible orthologue of a histone deacetylase Sir2-like protein (z-score of 2.5). Likewise, Tb927.11.10550 is a nuclear (and cytoplasmic) protein (Billington et al., Reference Billington, Halliday, Madden, Dyer, Barker, Moreira-Leite, Carrington, Vaughan, Hertz-Fowler, Dean, Sunter, Wheeler and Gull2023) that exhibits low sequence similarity to PF0610 (HHpred probability of 35%, E-value of 21), an archaeal protein presumably involved in transcription regulation (Wang et al., Reference Wang, Lee, Sugar, Jenney, Adams and Prestegard2007), to the α subunit of TFIIE (HHpred probability of 20%, E-value of 59), and a subunit of the GATOR complex (z-score of 15.5), an upstream regulator of the mTORC1 pathway, which controls RNAP III transcription (Loissell-Baltazar and Dokudovskaya, Reference Loissell-Baltazar and Dokudovskaya2021). In yeast, TORC1-dependent SUMOylation of C82 is required for RNAP III assembly and robust tRNA transcription (Chymkowitch et al., Reference Chymkowitch, Nguéa, Aanes, Robertson, Klungland and Enserink2017). In this regard, among the proteins that copurified with TbC82 we found a SUMO-interacting motif-containing protein (Table 1), suggesting that C82 could also be SUMOylated in trypanosomatids.
With LmC82 also copurified a protein related to TIP120 (also known as CAND1) (HHpred probability of 100%, E-value of 1.4 × 10−30), which has been implicated in transcription activation of all 3 nuclear RNAPs in vertebrates (Makino et al., Reference Makino, Yogosawa, Kayukawa, Coin, Egly, Wang, Roeder, Yamamoto, Muramatsu and Tamura1999). We also found an NLI-interacting factor-like phosphatase (Table 2) that is related to Fcp1, responsible for removing phosphates from the carboxy-terminal domain of the largest subunit of RNAP II (Hausmann and Shuman, Reference Hausmann and Shuman2003). Moreover, we identified LmjF.11.0950, a Leishmania-specific hypothetical protein that shows weak homology to mTERF-2 (HHpred probability of 42%, E-value of 30), to the SPT16 subunit of the FACT complex (HHpred probability of 23%, E-value of 100), and the alpha subunit of CTD kinase (z-score of 4.5).
In yeast, tRNA genes associate with nuclear pore complexes to coordinate RNAP III transcription with the nuclear export of tRNAs (Chen and Gartenberg, Reference Chen and Gartenberg2014). In that respect, several components of the nuclear pore complexes were identified with LmC82, such as nucleoporin NUP96, importin 1 and a putative transportin 1. Thus, as in yeast, RNAP III complexes might bind to nuclear pore proteins in L. major. Of note, no nuclear pore proteins were identified with TbC82. Among the RNA-binding proteins that copurified with LmC82, we found PUF1, involved in fine-tuning gene expression in trypanosomatids (Luu et al., Reference Luu, Brems, Hoheisel, Burchmore, Guilbride and Clayton2006), as well as the La protein, which not only binds to RNA molecules transcribed by RNAP III, but also to mRNAs and other transcripts (Sommer and Heise, Reference Sommer and Heise2021). Actin copurified with both TbC82 and LmC82 (Tables 1 and 2). While it is a frequent contaminant in tandem affinity purifications, it has been demonstrated that human actin interacts with RNAP III to promote the synthesis of the U6 snRNA in a reconstituted in vitro transcription system (Hu et al., Reference Hu, Wu and Hernandez2004). Therefore, actin may interact with C82 in trypanosomatids to control RNAP III activity.
It is important to point out that the C82-PTP protein was produced from an episome in L. major, which could result in the overproduction of the protein. In contrast, the C82 gene was PTP-tagged in situ in T. brucei. Nevertheless, previous works have shown comparable results with these 2 different approaches. For instance, the RNA editing complexes isolated in L. tarentolae (Aphasizhev et al., Reference Aphasizhev, Aphasizheva, Nelson, Gao, Simpson, Kang, Falick, Sbicego and Simpson2003) by overexpressing TAP-tagged proteins are very similar to those identified in T. brucei by in situ tagging (Schnaufer et al., Reference Schnaufer, Ernst, Palazzo, O'Rear, Salavati and Stuart2003). Also, similar base J binding protein complexes have been reported in L. major (by overexpressing TAP-tagged proteins) (Jensen et al., Reference Jensen, Phan, McDonald, Sur, Gillespie, Ranish, Parsons and Myler2021) and T. brucei (Kieft et al., Reference Kieft, Zhang, Marand, Moran, Bridger, Wells, Schmitz and Sabatini2020). Moreover, comparable CTR9 complexes were isolated in L. major (by episomal overexpression of tagged protein) (Jensen et al., Reference Jensen, Phan, McDonald, Sur, Gillespie, Ranish, Parsons and Myler2021) and T. brucei (by in situ tagging) (Ouna et al., Reference Ouna, Nyambega, Manful, Helbig, Males, Fadda and Clayton2012). Thus, while overexpression of tagged proteins in Leishmania might potentially lead to the association with contaminants, the majority of the most abundant proteins represent real interactors; and non-specific proteins are also copurified in T. brucei by in situ tagging. Consequently, we believe that the differences we observed between L. major and T. brucei are genuine, and not the result of using different approaches to express the tagged C82 proteins.
In conclusion, in this work we have demonstrated that both TbC82 and LmC82 localize to the nucleus, where they associate in vivo with 5S rRNA, tRNA and U2 snRNA genes. Ablation of TbC82 led to a strong reduction in the levels of RNAP III-dependent transcripts and to the death of insect forms of T. brucei. The 2 isoforms of C82 present in L. major are expressed at the protein level and seem to be present in RNAP III complexes. Multiple putative TbC82 and LmC82 interactors were identified, including RNAP subunits such as C34. However, the orthologue of subunit C31 was not found in these parasites. Notably, our results suggest a robust association of C82 with TFIIIC in T. brucei, but not in L. major. Likewise, several components of the nuclear pore complexes copurified exclusively with LmC82. Moreover, novel putative regulators of transcription were identified in both parasites, some of which are genus-specific. Thus, our results indicate that RNAP III complexes are not identical in both parasites. The observed differences in the predicted 3-dimensional structures of TbC82 and LmC82 may be related, at least in part, to the distinctive interactions that they establish, which added to the presence of 2 C82 isoforms in L. major, most likely reflect differences in RNAP III transcription regulation between T. brucei and L. major. Interestingly, human cells possess 2 isoforms of subunit C31 (RPC7α and RPC7β) that give rise to 2 forms of RNAP III (RNAP IIIA and RNAP IIIB). Differential expression of these 2 isoforms (and the RNAP III forms) has been observed during early embryogenesis, in differentiated cells, and during tumorigenesis (Cheng and Van Bortle, Reference Cheng and Van Bortle2023). It would be interesting to explore whether different forms of RNAP III exist in L. major.
Several differences in the transcription process have been reported between T. brucei and L. major. For instance, RNAP I transcribes several protein-coding genes in T. brucei, but not in Leishmania (Günzl et al., Reference Günzl, Bruderer, Laufer, Schimanski, Tu, Chung, Lee and Lee2003). Also, base J in Leishmania is required for proper transcription termination throughout the genome, but in T. brucei base J does not regulate transcription termination at most convergent SSRs (Reynolds et al., Reference Reynolds, Cliffe, Förstner, Hon, Siegel and Sabatini2014). Moreover, retrotransposon hot spot proteins are trypanosome-specific factors (not found in Leishmania) that interact with RNAP II, and whose depletion impairs mRNA synthesis (Florini et al., Reference Florini, Naguleswaran, Gharib, Bringaud and Roditi2019). Also, while the tRNA-Sec is transcribed by RNAP II in T. brucei (Aeby et al., 2010), it is synthesized by both RNAP II and RNAP III in L. major (Padilla-Mejía et al., Reference Padilla-Mejía, Florencio-Martínez, Moreno-Campos, Vizuet-de-Rueda, Cevallos, Hernández-Rivas, Manning-Cela and Martínez-Calvillo2015).
Supplementary material
The supplementary material for this article can be found at https://doi.org/10.1017/S0031182024000921.
Data availability statement
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository (https://www.ebi.ac.uk/pride/) with the dataset identifiers PXD051017 and 10.6019/PXD051017.
Acknowledgements
This work is one of the requirements to obtain the PhD degree in Posgrado en Ciencias Biomédicas, Universidad Nacional Autónoma de México (UNAM), for José Andrés Cano Santiago, who was the recipient of doctoral fellowship 916313 from CONAHCYT. We also thank Raúl Bobes-Ruíz and Abraham Landa-Piedra for fruitful discussions, and Ebbing De Jong (Core Facility for Proteomics and Mass Spectrometry from Upstate Medical University, NIH shared instrumentation grant 1S10OD023617–01A1) for mass spectrometry analyses.
Author contributions
A. C.-S.: conducted experiments, in silico analysis, data analysis, writing (draft preparation). L. E. F.-M.: conducted experiments, figure preparation. D. E. V.-R.: conducted experiments. A. J. R.-C.: conducted experiments. R. G. M.-C.: resources, writing (review and editing). T. N.-M.: in silico analysis, figure preparation, funding acquisition, writing (review and editing). S. M.-C.: conceptualization, funding acquisition, supervision, data analysis, writing (original draft preparation, review and editing).
Financial support
This work was supported by grants IN214221 and IN208224 from UNAM-PAPIIT, and grant CF-2023-I-820 from CONAHCYT to S. Martínez-Calvillo; and by grants IA200623 and IA204721 from UNAM-PAPIIT to T. Nepomuceno-Mejía.
Competing interests
None.
Ethical standards
All applicable international, national, and institutional guidelines for the care and use of mice were followed.