Introduction
Cystic echinococcosis (CE) is a zoonotic infectious disease caused by several members of the cestode species complex Echinococcus granulosus sensu lato (s.l.). CE is considered by the World Health Organization as one of the most important ‘neglected zoonotic diseases’ (WHO, 2020). The life cycle of these parasites is obligate between carnivorous definitive (mostly canids) and herbivorous intermediate hosts (Romig et al., Reference Romig, Deplazes, Jenkins, Giraudoux, Massolo, Craig, Wassermann, Takahashi and Rue2017). The adult worms, which are only a few millimetres in length, live in the small intestine of the definitive hosts, and their eggs are released into the environment with the feces. After oral ingestion of the eggs via contaminated water or food by intermediate hosts, the larval cyst-like metacestode may develop in various organs, most often in the liver and lungs. The protoscoleces, which develop in large numbers in these cysts, grow into adult worms when ingested by the definitive host during predation or scavenging (Thompson, Reference Thompson2017). If infectious eggs are accidentally ingested by humans, development of metacestodes in internal organs may occur, leading to CE due to damage of organs by the growth of cysts, which may ultimately be fatal (Kern et al., Reference Kern, da Silva, Akhan, Müllhaupt, Vizcaychipi, Budke and Vuitton2017; Thompson, Reference Thompson2017).
The species complex E. granulosus s.l. consists of 5 distinct species: E. granulosus sensu stricto (s.s.), Echinococcus equinus, Echinococcus ortleppi, Echinococcus felidis and Echinococcus canadensis. The latter species is the most diverse, holding distinct genotypes (G6, G7, G8 and G10), and there is debate whether E. canadensis should be split into 2 or even 3 species (Lymbery et al., Reference Lymbery, Jenkins, Schurer and Thompson2015; Yanagida et al., Reference Yanagida, Lavikainen, Hoberg, Konyaev, Ito, Sato, Zaikov, Beckmen and Nakao2017; Laurimäe et al., Reference Laurimäe, Kinkar, Moks, Romig, Omer, Casulli, Umhang, Bagrade, Irshadullah, Sharbatkhori, Mirhendi, Ponce-Gordo, Soriano, Varcasia, Rostami-Nejad, Andresiuk and Saarma2018a). The various genotypes, formerly named ‘strains’, differ with respect to life cycle, geography and genetics. Although the closely related and globally distributed G6 and G7 are mostly associated with a livestock–dog life cycles (often involving camels or pigs), G8 and G10 only occur in the Northern Hemisphere and are sylvatic in nature involving mainly wolves and cervids such as moose and reindeer as hosts (Romig et al., Reference Romig, Deplazes, Jenkins, Giraudoux, Massolo, Craig, Wassermann, Takahashi and Rue2017). According to the distribution of their hosts, G8 and G10 can be found in the northern temperate to Arctic latitudes (Romig et al., Reference Romig, Deplazes, Jenkins, Giraudoux, Massolo, Craig, Wassermann, Takahashi and Rue2017).
While all variants of G6 and G7 are genetically close (and are often referred to as the G6/7 cluster) (Addy et al., Reference Addy, Wassermann, Kagendo, Ebi, Zeyhle, Elmahdi, Umhang, Casulli, Harandi, Aschenborn, Kern, Mackenstedt and Romig2017), the relationship of G8 and G10 with each other and with G6/7 is differently resolved depending on whether nuclear or mitochondrial DNA is considered. Analysis of the mitochondrial genome suggests a closer affinity of G10 to G6/7, while some nuclear marker genes support a clade with G8 and G10 (Yanagida et al., Reference Yanagida, Lavikainen, Hoberg, Konyaev, Ito, Sato, Zaikov, Beckmen and Nakao2017; Laurimäe et al., Reference Laurimäe, Kinkar, Moks, Romig, Omer, Casulli, Umhang, Bagrade, Irshadullah, Sharbatkhori, Mirhendi, Ponce-Gordo, Soriano, Varcasia, Rostami-Nejad, Andresiuk and Saarma2018a).
Taxonomic decisions on E. canadensis have been postponed so far due to a lack of sufficient data, especially on the northern genotypes G8 and G10. In contrast to G6/7, whose genetic structure is well known, only very few specimens of G8 and G10 have ever been characterized and showed little intra-strain variability. However, the great majority came from the European region, and the study of the nuclear genome was performed exclusively with sample material from Europe (Laurimäe et al., Reference Laurimäe, Kinkar, Moks, Romig, Omer, Casulli, Umhang, Bagrade, Irshadullah, Sharbatkhori, Mirhendi, Ponce-Gordo, Soriano, Varcasia, Rostami-Nejad, Andresiuk and Saarma2018a). The northeastern part of Asia is particularly data-deficient, although this is the only known region where all genotypes of E. canadensis have been recorded, leading to speculation on the geographic origin of the species (Konyaev et al., Reference Konyaev, Yanagida, Nakao, Ingovatova, Shoykhet, Bondarev, Odnokurtsev, Loskutova, Lukmanova, Dokuchaev, Spiridonov, Alshinecky, Sivkova, Andreyanov, Abramov, Krivopalov, Karpenko, Lopatina, Dupal, Sako and Ito2013; Ito et al., Reference Ito, Dorjsuren, Davaasuren, Yanagida, Sako, Nakaya, Nakao, Bat-Ochir, Ayushkhuu, Bazarragchaa, Gonchigsengee, Li, Agvaandaram, Davaajav, Boldbaatar and Chuluunbaatar2014; Zhang et al., Reference Zhang, Yang, Zeng, Zhao, Liu, Piao, Jiang, Cao, Shen, Liu and Zhang2014; Yang et al., Reference Yang, Zhang, Zeng, Zhao, Zhang and Liu2015; Wu et al., Reference Wu, Li, Zhu, Li, Zhang, Li, Yao, Tian, Fu, Yin, Zhu, HongBin and WanZhong2018; Hua et al., Reference Hua, Xie, Song, Shi, Zhan, Wu, Gu, Peng and Yang2019).
In the present study, we collected samples of Echinococcus spp. from a variety of wild and semi-domestic definitive and intermediate hosts in the Republic of Sakha (Russian Federation), and examined them for species, genotypes and haplotypic diversity to obtain new insights into the structure of the E. canadensis cluster.
Materials and methods
Parasite material
The parasite material originated from wild and semi-domestic animals from the Republic of Sakha, located in the Russian Far East in northeast Asia (Fig. 1).

Figure 1. Map of Russia with the Republic of Sakha (red), blue ellipse shows area of sample collection in Sakha; green ellipse shows Finland (source: www.wikipedia.com; CC BY-SA 4.0).
Echinococcus worms from definitive hosts and cyst material from intermediate hosts were available for the present study. Adult worms were isolated from small intestines of 94 legally hunted wolves (Canis lupus) from Sakha in the period 2011–2015 and preserved in a fixative solution. One cyst each from moose (Alces alces), elk (Cervus canadensis) and roe deer (Capreolus pygargus) and 2 from reindeer (Rangifer tarandus) could be opportunistically collected from hunted animals in Sakha during meat inspection and preserved in 70% ethanol (EtOH).
In addition, 13 cyst samples of E. canadensis G10 stored in 70% EtOH from 3 reindeer and 10 moose from Finland were also available for molecular analyses (Fig. 1).
Sample preparation
The content of the sample tubes with the adult worm material was examined under an inverted microscope. Individual worms or worm fragments were aspirated with a pipette and each transferred to a 200 μL polymerase chain reaction (PCR) tube containing 20 μL of 0.02 m sodium hydroxide (NaOH). Of the cyst samples, a small tissue piece of ~1 × 1 mm2 in size was cut with a scalpel and each transferred to separate tubes containing 30 μL of 0.02 m NaOH. The isolated single worms and cyst material were lysed at 95°C for 15 min (Nakao et al., Reference Nakao, Sako and Ito2003). The lysate was used directly as a template for the subsequent molecular analyses or stored at −20°C until further use. For the cyst samples that failed to yield a positive PCR result, DNA was obtained by proteinase K digestion followed by phenol–chloroform extraction and EtOH precipitation as described previously (Dinkel et al., Reference Dinkel, von Nickisch-Rosenegk, Bilger, Merli, Lucius and Romig1998).
DNA amplification and sequencing
Species identification was done by nested PCR and sequencing. The target gene was the ~1600 bp long mitochondrial cytochrome c oxidase subunit 1 (cox1) gene. To obtain the complete cox1 gene, different primer combinations were used targeting the front part or the back part of the gene. If the PCR remained negative, an attempt was made to amplify a cox1 fragment of ~200 bp in length to identify the species. If this attempt also remained negative, additional fragments of other mitochondrial genes, the cytochrome b (cob, ~170 bp) or NADH dehydrogenase subunit 1 gene (nad1, ~190 bp), were targeted. All primers used are listed in Table 1.
Table 1. Primer pairs used for PCR

a Hüttner et al. (Reference Hüttner, Nakao, Wassermann, Siefert, Boomker, Dinkel, Sako, Mackenstedt, Romig and Ito2008).
F, forward primer; R, reverse primer.
The amplification was done via nested PCR. For the first PCR, a reaction volume of 25 μL was prepared. The mixture contained 10 mm Tris-hydrochloric acid (Tris-HCl) (pH 8.3), 50 mm potassium chloride (KCl), 2 mm magnesium chloride (MgCl2), 200 μM of each deoxynucleotide triphosphates (dNTPs), 6.25 pmol of each first PCR primer and 0.625 U of Taq polymerase (Applied Biosystems, Carlsbad, CA, USA). One microlitre of the cyst or worm lysate, or extracted DNA was added to the PCR mixture as a template. The volume of the nested PCR reaction mixture was 50 μL and consisted of 10 mm Tris-HCl (pH 8.3), 50 mm KCl, 2 mm MgCl2, 200 μ m of each dNTPs, 12.5 pmol of each nested PCR primer and 1.25 U of Taq polymerase and 2 μL of the first PCR product as template DNA.
The conditions during amplification for the PCRs were as follows: initial denaturation at 95°C for 5 min followed by 35 cycles with denaturation at 95°C for 30 s, annealing at 50°C for 30 s, elongation at 72°C for 60 s (for front and back parts of cox1 gene) or 30 s (for small fragments of cox1, cob and nad1 genes) and a final elongation at 72°C for 5 min. Amplification results were obtained on a 1.5% agarose gel stained with GelRed™. PCR products were purified with a High Pure PCR Product Purification Kit (Roche, Mannheim, Germany) and sent to Microsynth Seqlab GmbH (Göttingen, Germany) for sequencing. The sequences were viewed, edited and assembled using the GENtle V1.9.4 program (Manske M., University of Cologne, Germany) and compared with GenBank entries using the NCBI basic local alignment search tool (BLAST) for species identification.
Analyses of haplotype networks and diversity indices
The haplotypes and the genetic diversity of the obtained complete cox1 sequences were analysed. Haplotype identification and creation of haplotype networks were calculated with TCS 1.2 software (Clement et al., Reference Clement, Posada and Crandall2000) using statistical parsimony (Templeton et al., Reference Templeton, Crandall and Sing1992). Networks were visualized using the online program tcsBU (Santos et al., Reference Santos, Cabezas, Tavares, Xavier and Branco2016). The haplotype and nucleotide diversity indices were analysed by DnaSP 6.12 (Rozas et al., Reference Rozas, Ferrer-Mata, Sánchez-DelBarrio, Guirao-Rico, Librado, Ramos-Onsins and Sánchez-Gracia2017) and analysis of molecular variance, degree of genetic differentiation (pairwise fixation index, Fst) and neutrality indices of Tajima's D and Fu's F s were calculated using Arlequin v 3.5.2.2 software (Excoffier et al., Reference Excoffier, Laval and Schneider2005).
In addition to 13 samples from Finland examined here, all E. canadensis G10 sequences of the complete cox1 gene available in GenBank were included in the analyses for comparison. The samples behind the sequences originated from eastern Russia (Sakha), Mongolia, China (Tibet), the USA (Alaska) and Europe (Estonia, Sweden, Finland and Russia) and are listed in Table 2. For the comparison of the genetic diversity and neutrality indices of the sequences found in northeast Asia and Europe, the frequencies of the haplotypes discovered in different studies were extracted from the corresponding literature (Table 2).
Table 2. Geographic origin, host, haplotype, accession number and reference of samples and sequences used for the analyses

a Origin given in Nakao et al. (Reference Nakao, Yanagida, Konyaev, Lavikainen, Odnokurtsev, Zaikov and Ito2013) as ‘Far East Russia’, specified in Konyaev et al. (Reference Konyaev, Yanagida, Nakao, Ingovatova, Shoykhet, Bondarev, Odnokurtsev, Loskutova, Lukmanova, Dokuchaev, Spiridonov, Alshinecky, Sivkova, Andreyanov, Abramov, Krivopalov, Karpenko, Lopatina, Dupal, Sako and Ito2013) as ‘Yakutia’.
Results
Parasite species identification
Adult worms or worm fragments could be isolated from the 94 wolf samples. Depending on the sample, between 1 and 36 worms or worm fragments, in total 713 were isolated individually and examined. Amplification of at least one of the small 170–200 bp gene fragments was successful only in 191 worms from 35 samples. Echinococcus canadensis G10 was detected in 15 wolves (1–14 worms per wolf) and Echinococcus multilocularis in 2 wolves (1 and 2 worms). Of the E. multilocularis specimens only the small nad1 fragment could be amplified resulting in ~100 bp long sequences. The comparison with GenBank did not allow an assignment to known haplotypes. Identical sequences were detected in isolates from e.g. Europe, China or St. Lawrence Island. Twenty-one wolves showed infection with juvenile Taenia sp., of which a ~200 bp nad1 fragment had 95.5% similarity with Taenia multiceps (NC012894; Jia et al., Reference Jia, Yan, Guo, Zhu, Wang, Shi, Chen, Zhan, Zhang, Fu, Littlewood and Cai2010); 2 of these wolves were co-infected with E. canadensis G10. In 38 E. canadensis G10 worms from 10 wolves, amplification and sequencing of the complete cox1 gene was successful. Analysis of the intermediate hosts also revealed infection with E. canadensis G10 in 2 reindeer, 1 moose and 1 elk; the cyst from the roe deer could not be amplified. All of the 13 cyst samples from Finland (3 reindeer and 10 moose) belonged to E. canadensis G10. From all these cyst samples the complete cox1 gene could be sequenced. A total of 42 cox1 sequences from Sakha and 13 sequences from Finland were thus available for the further haplotype analysis (Table 2). The sequences were deposited at NCBI GenBank under the accession numbers OR420689–OR420703.
Number of haplotypes and parsimony network
A total of 99 complete cox1 gene sequences of E. canadensis G10, including the present sequences and GenBank entries, were available for the calculation of number of haplotypes and network.
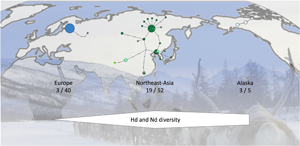
Analysis of the sequences of the 42 samples from Sakha revealed 15 haplotypes of E. canadensis G10, while only 1 haplotype was detected in the Finnish samples of 13 cysts (Table 2). When all 99 sequences were analysed according to their geographic origin, 3 haplotypes could be detected in 40 sequences from Europe, 17 haplotypes in the 47 eastern Russian sequences (42 from the present study and 5 additional from GenBank) and 19 haplotypes in northeast Asia (including the sequences from eastern Russia, and GenBank entries from Mongolia and Tibet). The 5 sequences with Alaskan origin resulted in 3 haplotypes.
In addition to the 99 E. canadensis G10 sequences, 1 sequence each of genotypes 6, 7 and 8 of E. canadensis was included in the haplotype network (Fig. 2, Table 2). The network has 3 major haplotypes. Two are found in northeast Asia (H01, H02) and the third is the dominant haplotype in Europe (H19), comprising all but 2 of the 40 European sequences. The sequences of 2 intermediate hosts from Sakha were identical to the European major haplotype and 1 sequence of European origin was closely located to the haplotypes from northeast Asia and identical to one from eastern Russia (H05). Apart from these 2 haplotypes, there were no further shared haplotypes between Europe and northeast Asia. The 3 Alaskan variants were clearly separated from the other haplotypes and formed a separate branch (Fig. 2).

Figure 2. Haplotype network of Echinococcus canadensis G10 based on complete cox1 gene (1608 bp). Sequences detected in Sakha in the present study are shown in dark green. The size of the circles indicating the frequencies of haplotypes, small colourless circles showing hypothetical haplotypes (hh). E. can = E. canadensis.
A total of 15 haplotypes of E. canadensis G10 were detected in 10 wolves; this means that in some wolves several haplotypes were identified. Between 1 and 6 worms could be analysed per animal and in some wolves all worms possessed the same cox1 gene variant (e.g. 6 worms from wolf 4 belonging to haplotype H01), in others all worms showed a different haplotype (e.g. 5 worms each from wolf 7 and 8) (Table 2).
Population diversity and neutrality indices
Analyses of the haplotype (H d) and nucleotide diversity (N d) indices show low values in Europe, compared to the diversity found in northeast Asia, which are 8.3 (H d) and 12.1 (N d) times higher, respectively (Table 3). The value of European diversity H d was 0.0987 and that of nucleotide diversity N d was 0.0078. The indices from Sakha determined in the present study are almost 1 order of magnitude higher compared to Europe (H d: 0.7526; N d: 0.0694) and increase further with the inclusion of the other sequences from eastern Russia, Mongolia and China (H d: 0.8204; N d: 0.0943). The Alaskan samples show also high haplotype diversity values (H d: 0.8000); the nucleotide diversity (N d: 0.0313) is lower than that in northeast Asia, but still higher compared to Europe.
Table 3. Diversity and neutrality indices of E. canadensis G10 based on complete cox1 gene (1608 bp)

n, number of isolates (sequences); n H, number of haplotypes; H d, haplotype diversity; N d, nucleotide diversity; s.d., standard deviation.
a Including sequences from Sakha and additional from eastern Russia.
b Including sequences from eastern Russia, Mongolia, and Tibet.
* Significant at P < 0.05.
Statistically significant negative Tajima's D values were found in all populations except for Alaska, which had a positive value, but this should be interpreted with caution due to the small number of samples. The Fu's F s values were all negative, but only the northeast Asian populations were significant (Table 3).
Population differentiation (F ST)
The fixation index (F ST) was calculated to estimate the population differentiation between the European, the different groups of northeast Asian and Alaskan sequences. The F ST values between the European population and all others were significantly high, similar to between Alaska and northeast Asia, which means that the populations are quite separate and there is little genetic exchange (Table 4). In comparison, the F ST of Alaska and northeast Asia is considerably smaller at 0.6875, but still represents a clear differentiation. The negative values when comparing the Sakha population with total eastern Russia or total northeast Asia can be considered as zero and show no genetic differentiation.
Table 4. F ST values between E. canadensis G10 populations based on complete cox1 gene (1608 bp)

a Including sequences from Sakha and additional from eastern Russia.
b Including sequences from eastern Russia, Mongolia and Tibet.
* Significant at P < 0.01.
Discussion
The taxonomic status of the 4 genotypes G6/7, G8 and G10, provisionally grouped under the name Echinococcus canadensis (Vuitton et al., Reference Vuitton, McManus, Rogan, Romig, Gottstein, Naidich, Tuxun, Wen and da Silva2020) the phylogenetically youngest Echinococcus species, is not yet fully resolved. As mentioned above, differences can be seen between the genotypes with respect to life cycle, geography and genetics (Romig et al., Reference Romig, Deplazes, Jenkins, Giraudoux, Massolo, Craig, Wassermann, Takahashi and Rue2017). The genotypes 6/7 involve primarily domesticated animals in its cycle, whereas G8 and G10 both are linked to transmission by wild animals (Oksanen and Lavikainen, Reference Oksanen and Lavikainen2015; Romig et al., Reference Romig, Deplazes, Jenkins, Giraudoux, Massolo, Craig, Wassermann, Takahashi and Rue2017). Compared to G6/7, only a few studies were carried out with genotypes 8 and 10, and these only with small sample sizes as acquisition of samples from wild animals is difficult and therefore only a few samples are usually available for evaluation. Even more importantly, the majority of these specimens came from northern Europe, very few from northeast Asia and Alaska. A recent study investigated the genetic diversity of European specimens of E. canadensis G8 and G10 using the whole mitochondrial genome and found only a very small number of genetic variants within the 2 genotypes (3 haplotypes among 14 G8 samples and 5 among 15 G10 samples) (Laurimäe et al., Reference Laurimäe, Kinkar, Moks, Bagrade and Saarma2023). All varied only in 1–3 nucleotide exchanges, except for 1 G10 isolate from Estonia, which had a higher variation with 24 exchanges. Considering that this variation occurred in the entire mitochondrial genome with more than 13 500 bp, the numbers of exchanges are small compared to the results of the Asian samples in the present study. The variation of the cox1 gene (1608 bp) alone is thus remarkable and unexpected. The largest distance between the haplotypes found in Sakha was 8 nucleotide exchanges. In total, i.e. the 42 sequences analysed here and the 5 additional sequences from the GenBank, which also originate from eastern Russia, 17 gene variants could be detected. Including the sequences from China and Mongolia, 19 haplotypes can be found in 54 northeast Asian sequences. The European cox1 sequences showed only 3 haplotypes, whereby the above-mentioned Estonian sequence was the most distant with a difference of 4 bases to the major European haplotype. Interestingly, this gene variant was also described from the very east of Sakha (Konyaev et al., Reference Konyaev, Yanagida, Nakao, Ingovatova, Shoykhet, Bondarev, Odnokurtsev, Loskutova, Lukmanova, Dokuchaev, Spiridonov, Alshinecky, Sivkova, Andreyanov, Abramov, Krivopalov, Karpenko, Lopatina, Dupal, Sako and Ito2013); and conversely, the European main variant was also found in 2 intermediate hosts from Sakha. Even as this indicates some minor genetic exchange between Europe and eastern Asia, it concerns only 3 out of 99 sequences and is therefore at an extremely low level.
In general, there is a surprisingly high genetic variation of G10 in northeast Asia. Even more so when one considers that the 38 worms from the present study came from only 10 wolves. In some wolves, all the worms analysed had the same haplotype, but 6 wolves possessed worms with different gene variants. In 2 cases, each of the 5 isolated worms showed a different haplotype. This suggests a wide distribution and high prevalence of the parasite in the intermediate hosts in this region.
Unfortunately, we could not obtain the complete cox1 sequence from the remaining isolates despite multiple repetitions. In those cases, DNA isolation using NaOH was followed by the classical digestion and the phenol–chloroform extraction method as well as a commercial extraction kit. In none of the cases could an amplification product be obtained afterwards, even with large Taenia worms (results not shown), so it is assumed that the DNA was degraded during storage for unknown reasons. This prevented also the analyses of further genes of the samples with complete cox1 sequence, since multiple repetitions were needed to obtain this single gene and DNA solutions were depleted in most cases of adult worms. The analyses of other mitochondrial and in particular nuclear genes would have been desirable, as this could have helped to clarify the question of whether it could be justified to split E. canadensis into 2 or 3 species.
The results of the present study show that the genetic diversity of E. canadensis G10 in Northeast Asia is far higher than elsewhere, particularly when compared to northern Europe. Intriguingly, northeast Asia is also the only region where all 4 genotypes of E. canadensis are known to occur. The northernmost identification of a G6 infection was done in Sakha by Konyaev et al. (Reference Konyaev, Yanagida, Nakao, Ingovatova, Shoykhet, Bondarev, Odnokurtsev, Loskutova, Lukmanova, Dokuchaev, Spiridonov, Alshinecky, Sivkova, Andreyanov, Abramov, Krivopalov, Karpenko, Lopatina, Dupal, Sako and Ito2013) who found a reindeer infected with this genotype. Further south, in Mongolia and the Tibetan Plateau of China, E. canadensis G6/7 is common and G10 has also been identified (Ito et al., Reference Ito, Dorjsuren, Davaasuren, Yanagida, Sako, Nakaya, Nakao, Bat-Ochir, Ayushkhuu, Bazarragchaa, Gonchigsengee, Li, Agvaandaram, Davaajav, Boldbaatar and Chuluunbaatar2014; Zhang et al., Reference Zhang, Yang, Zeng, Zhao, Liu, Piao, Jiang, Cao, Shen, Liu and Zhang2014; Yang et al., Reference Yang, Zhang, Zeng, Zhao, Zhang and Liu2015; Wu et al., Reference Wu, Li, Zhu, Li, Zhang, Li, Yao, Tian, Fu, Yin, Zhu, HongBin and WanZhong2018). The Tibetan Plateau is the southernmost area where G8 and G10 can still be found in addition to G6/7 (Wu et al., Reference Wu, Li, Zhu, Li, Zhang, Li, Yao, Tian, Fu, Yin, Zhu, HongBin and WanZhong2018; Hua et al., Reference Hua, Xie, Song, Shi, Zhan, Wu, Gu, Peng and Yang2019). Despite this diversity, our data indicate that E. canadensis genotypes are not randomly scattered throughout the region, as only G10 was detected among 191 amplifiable worms from 35 wolves, and cysts from 4 cervids in Sakha, while not a single isolate of G8 could be found. We can only speculate about the reasons. In Europe, records of G8 are mainly from more temperate regions, with G10 venturing further north (Laurimäe et al., Reference Laurimäe, Kinkar, Moks, Bagrade and Saarma2023). Our study area in Sakha boasts some of the lowest temperature records outside Antarctica, with winter temperatures that may fall below −60°C. This certainly puts some stress on a parasite life cycle that includes environmental stages (eggs), but whether G10 is better adapted to such conditions than other genotypes is open to further investigations.
Two of the wolves were infected with E. multilocularis, indicating that wolves may be involved in the life cycle of this parasite in Sakha as it has been shown in the subarctic parts of North America (Gesy et al., Reference Gesy, Schurer, Massolo, Liccioli, Elkin, Alisauskas and Jenkins2014).
We cannot draw any conclusion on the identity of the Taenia species that was present in 21 of the 35 wolves with amplifiable DNA. The sequence did not match any GenBank deposit, and only 95.5% identity rules out conspecificity with T. multiceps. The presence of only juvenile worms is likely due to sampling bias, as only Echinococcus-sized worms were targeted. Further investigations, also on cysticerci from wolf prey species, are required to identify this obviously cryptic species.
The exploration of the genetic diversity can shed light on the origin of species, as has been shown for E. granulosus s.s. Based on differences in haplotype diversity of E. granulosus s.s. from different regions, this parasite is thought to have originated in the Middle East, in the area of the Fertile Crescent. There, the genetic diversity of the parasite is highest, and it decreases with increasing geographical distance (Casulli et al., Reference Casulli, Interisano, Sreter, Chitimia, Kirkova, Rosa and Pozio2012; Yanagida et al., Reference Yanagida, Mohammadzadeh, Kamhawi, Nakao, Sadjjadi, Hijjawi, Hafez, Sako, Okamoto and Ito2012). As for E. canadensis, the genetic haplotype diversity has only been investigated from the G6/7 genotypic cluster. Although some geographical structuring can be seen with the ‘G6’ variants originating mostly from camel-raising regions and G6/7 contains a large number of haplotypes, most of these cluster closely together suggesting close relationship and recent ancestry (Addy et al., Reference Addy, Wassermann, Kagendo, Ebi, Zeyhle, Elmahdi, Umhang, Casulli, Harandi, Aschenborn, Kern, Mackenstedt and Romig2017; Laurimäe et al., Reference Laurimäe, Kinkar, Romig, Omer, Casulli, Umhang, Gasser, Jabbar, Sharbatkhori, Mirhendi, Ponce-Gordo, Lazzarini, Soriano, Varcasia, Rostami-Nejad, Andresiuk, Maravilla, González, Dybicz, Gawor, Šarkūnas, Šnábel, Kuzmina and Saarma2018b). In contrast, many haplotypes of G10 analysed in this study are widely separated from each other as reflected by the far higher value of nucleotide diversity (N d = 0.0943) as compared to G6/7 (N d ≤ 0.00173) (Addy et al., Reference Addy, Wassermann, Kagendo, Ebi, Zeyhle, Elmahdi, Umhang, Casulli, Harandi, Aschenborn, Kern, Mackenstedt and Romig2017).
The remarkably high genetic diversity of G10 in northeast Asia, the occurrence of all E. canadensis genotypes in this region and the phylogenetically basal position of genotypes 8 and 10 may be circumstantial evidence that this parasite might have evolved in this region.
However, more samples are needed from northern Russia, covering the vast area between Sakha and Europe, and from North America. More importance should be given not only to G10, but also to the analysis of the even more elusive G8, the most basally localized genotype. Assuming our tentative hypothesis that the geographic origin of the entire E. canadensis cluster is northeast Asia, a gradual decline in diversity towards the West and East (and to North America) should be observed.
Data availability statement
The authors confirm that the data supporting the findings of this study are available within the article.
Author's contribution
M. W., L. K. and T. R. conceived and designed the study. L. K., I. O. and A. O. conducted the sample collection. M. W., S. L. and J. O. carried out the laboratory analyses. M. W. and F. A. performed the computer analyses. All authors wrote and revised the manuscript.
Financial support
This study was funded by the Deutsche Forschungsgemeinschaft – DFG, Germany (grant number: WA-4336/1-1).
Competing interests
None.