INTRODUCTION
The increasing occurrence of clinical and especially subclinical Salmonella spp. infections in pig production worldwide, and the correlation between the within-herd prevalence and contamination of pig carcasses at slaughter [Reference De Busser1], has led to the development and implementation of several Salmonella control programmes and regulations in pig production worldwide. In addition, the importance of pork and pork products in the transmission of this pathogen to humans has been previously highlighted [2].
According to Sanchez et al. [Reference Sanchez3] prevalence of Salmonella infection at the farm and animal level in North America and Europe is around 59% and 17%, respectively. However, the prevalence of Salmonella shedding pigs on farm is often lower, with an average of 6·6% [Reference Pires, Funk and Bolin4]. In Argentina as well as in other South American countries little is known about the regional prevalence of Salmonella in pig herds [Reference Villamil, González and Ruiz5, Reference Parada6]. Nevertheless, both human sporadic cases and larger Salmonella foodborne outbreaks have been reported in the region [Reference Campos7].
The knowledge of the epidemiological factors driving disease distribution and persistence is a prerequisite for the design of appropriate control measures that, in the case of Salmonella, could eventually mitigate the risk of infection for the general public. In this context, spatial analytical tools can help to assess the distribution of a disease in the population in order to identify areas of higher risk and associated risk factors with a heterogeneous spatial distribution [Reference Benschop8, Reference Alvarez9]. In Argentina the central region is the main pig production area in the country, and includes 86% of all herds with more than 200 sows in the country (http://www.senasa.gov.ar/cadena-animal/porcinos/informacion/informes-y-estadisticas). However, there is little information on the prevalence and distribution of the different Salmonella serotypes in the region and the most significant factors associated with their presence. Although other studies have reported the importance of nutrition and management practices in the level of Salmonella infection in the herd [Reference Benschop8, Reference Hautekiet10], the applicability of those results to the regional production systems present in Argentina is currently unknown.
The objective of this study was to determine the prevalence and spatial distribution of Salmonella-infected herds in the central region of Argentina, and to identify the predominant serotypes and epidemiological factors associated with increased risk of infection.
MATERIALS AND METHODS
Study population
In order to determine the prevalence of infection in the region of study, the main pig-producing area in the country, 52 commercial farrow-to-finish pig farms with more than 200 sows each (min–max: 200–6400) were sampled between October 2011 and March 2013. This sample size was established considering the expected herd prevalence reported by Sanchez et al. [Reference Sanchez3] of 59%, with a 90% confidence level and 10% precision, and adjusting for a finite population [according to the national agency register there were 278 farms of >200 sows in the study region (Servicio Nacional de Sanidad y Calidad Agroalimentaria, Argentina)]. Farms were selected by intentional non-probabilistic sampling, and the number was completed by convenience sampling. In selected farms 30 pigs at slaughter age (22 weeks old) were sampled. The number of individual samples per farm was estimated to detect Salmonella shedders at an expected prevalence of 6·6% [Reference Pires, Funk and Bolin4] with a 90% confidence level. Individuals were selected purposively so that no more than 3–4 pigs per pen were included and animals with diarrhoea were included when present. Faecal samples (20 g) were collected from the rectum of the sampled individuals and refrigerated until processing within 48 h of collection.
Epidemiological information from each sampled herd was collected on the day of sampling using a semi-structured questionnaire, and included geographical coordinates, breeding herd size and genetic, hygienic-lock facility, and other herd management and potential risk factors. Data about the use of footbath, pig flow (all-in/all-out, continuous), presence of diarrhoea at the time of sampling and feed in the fattening facilities were also recorded.
Laboratory diagnostic tests
Faecal subsamples (10 g) were analysed for Salmonella isolation according to the ISO 6579 Annex D method, including negative and positive (S. Bredeney) controls. One or two colonies from the xylose lysine deoxycholate plate were confirmed as Salmonella using metabolic tests and invA detection by PCR according to Malorny et al. [Reference Malorny11]. Isolates from up to five animals per farm were serotyped according to the 9th edition of the White–Kauffmann–Le Minor scheme (Pasteur Institute, France).
Data analysis
Herds were classified as positive (at least one positive pig to Salmonella enterica) or negative. All the farms were geo-referenced and plotted using QGIS software version 2.12.0 [12]. Evidence of global spatial clustering of positive farms was assessed using the Cuzick–Edwards (CE) test for detection of global clustering [Reference Cuzick and Edwards13]. The test was run up to the 10th neighbourhood level using ClusterSeer software (TerraSeer Inc., USA). Additionally, local clustering of positive herds was assessed by means of the Bernoulli model of the spatial scan statistic [Reference Kulldorff14] using SaTScan software version 9.4.2 [Reference Kulldorff15].
In herds where epidemiological data was available and information about all the variables was complete (n = 43, 82·7%), bivariate associations between the herd-level factors and the Salmonella herd status (positive/negative) were assessed by Fisher's exact tests and analysis of variance (ANOVA) for categorical and continuous (herd size) variables, respectively. Results obtained from the spatial analysis (status of the closest neighbour) were also considered as potential covariates. Variables for which an association with P⩽0·2 was detected were included in a multivariable logistic regression model, where the outcome variable was the presence/absence of Salmonella in the herd. The model with the lowest Akaike's Information Criterion (AIC) was selected as the final model, and its goodness-of-fit was assessed using the Hosmer and Lemeshow test. A ROC analysis was also performed to evaluate model accuracy. Statistical analyses were performed using R software (R Development Core Team, 2015).
RESULTS
Descriptive results
Salmonella spp. were isolated from 22 out of the 52 farms sampled [herd prevalence 42·3%, 95% confidence interval (CI) 28·4–56·1]. When this value was stratified by farm size, herd prevalence was higher in larger farms (>1001 sows, 78%) than in small (200–500 animals, 37%) and medium (501–1000, 31%) farms, although these differences were not significant (ANOVA, P = 0·320). In five farms the target sample size could not be achieved due to lack of animals (n = 2) or samples damaged during transportation (n = 3), and four farms were Salmonella positive (only one with 23 samples was negative).
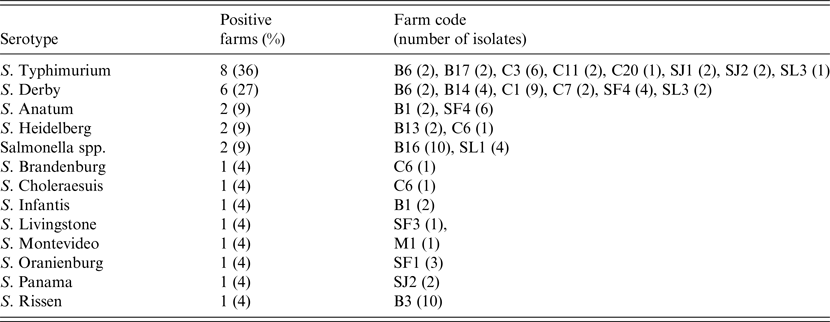
At the individual level 95 isolates were recovered from the 1518 faecal samples analysed, with a mean prevalence of shedder pigs, estimated in those herds classified as positive, of 13·6% (95% CI 8·0–19·2). Fifty-three of these isolates were serotyped, and S. Typhimurium and S. Derby were the most frequent serotypes, accounting for more than half of the positive farms (Table 1). A single serotype was identified in 59% of the positive farms, while in other farms two (22%) and three (9%) different serotypes were found (Table 1).
Table 1. Serotypes isolated in pigs in the 22 Salmonella-positive farms, including isolation frequency, farm identification code and number of isolates per farm
Spatial analysis
All the 22 Salmonella-positive herds were distributed throughout the study region (Fig. 1). The CE test detected significant level of clustering of the positive herds only at the first neighbourhood level (P = 0·01), while the Bernoulli model of the spatial scan statistic detected a cluster with borderline significance (P = 0·07) that included six farms in a radius of 73 km, with a relative risk of 2·88 (Fig. 1).
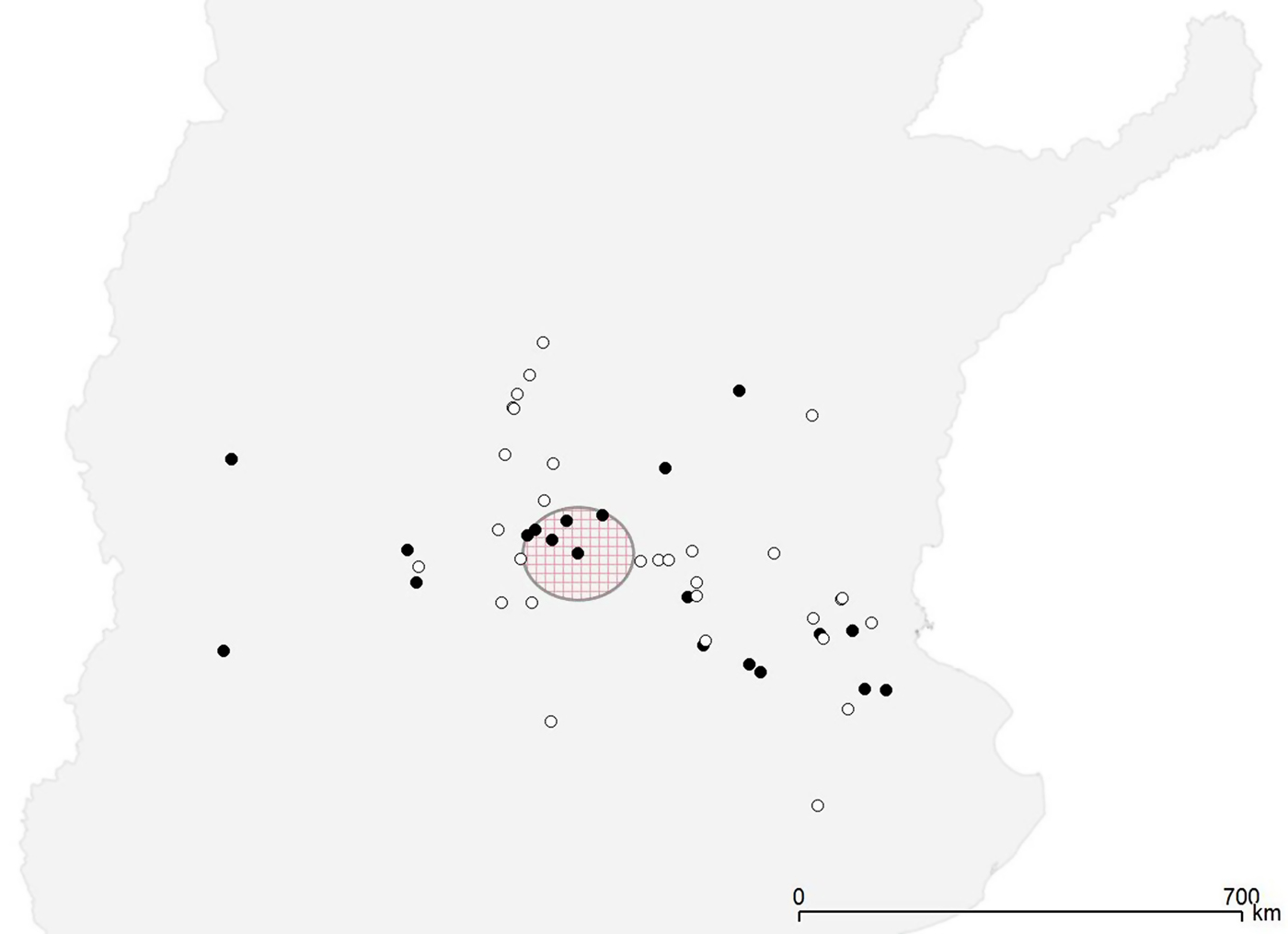
Fig. 1. Spatial distribution of Salmonella-positive (•) and Salmonella-negative (◦) farms. The large circle indicates the presence of a borderline significant (P = 0·08) cluster of high risk.
Epidemiological factor association
Presence of diarrhoea in finishing pigs and feed brand (as a global effect) were deemed potentially associated with the outcome of interest in the bivariate analysis (P < 0·2) and were thus included in the multivariate analysis. In addition, positive status of closest farm was also included due to the result in the bivariate analysis and in agreement with the result obtained in the CE test (Table 2). The final model with the lowest AIC included only the two first variables (P < 0·05), presence of diarrhoea in finishing pigs and feed brand: farms with diarrhoea in finishing pigs had significantly higher risk of harbouring positive animals [odds ratio (OR) 9·8, 95% CI 1·7–97·2], while farms using feed brand B were at significantly lower risk (OR 0·03, 95% CI 0·0–0·36) compared to brand A (Table 3).
Table 2. Point estimates and 95% confidence intervals (CI) of the odds ratios (OR) for positive results of bacteriology for Salmonella in pig herds (n = 43) from Argentina
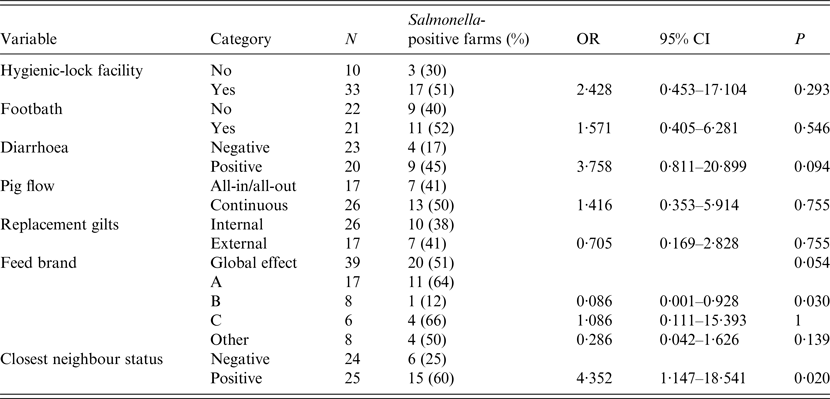
Table 3. Results of the multivariate model for positive results of Salmonella spp. at the farm level

OR, Odds ratio; CI, confidence interval.
The model had a good fit to the data as evidenced by the Hosmer–Lemeshow test (P = 0·86) and an adequate predictive capacity as demonstrated by the ROC analysis (area under the curve 81·4%).
DISCUSSION
The growing importance of the consumption of pork and pork product at a worldwide level highlights the need of a commitment of producers, veterinarians and other stakeholders involved in the production chain to accompany this growth with improved production systems. Incorporating new technologies and improving the nutrition and health of herds can help to ensure not only an increase in the number of pigs produced but also the food safety of their products. Despite agreement with previous studies, the risk of Salmonella contamination could be mitigated with different strategies at transport, lairage and along the slaughter line [Reference De Busser1], the assessment of the situation of this pathogen in the production systems is critical, not only for the obvious production losses resulting from possible medical conditions, but mainly because it is a major foodborne pathogen with a high impact on public health worldwide.
The results demonstrat that Salmonella was present in several herds throughout the study region. Moreover, the detection of spatial clustering at the first neighbourhood level suggest the existence of local transmission between herds or a potential exposure to a common source of infection. This would be in agreement with the results found in the spatial scan statistic, which identified a region in which farms were at a borderline significantly higher risk of testing positive for Salmonella (Fig. 1). According to Benschop et al. [Reference Benschop8], who also found a first-order spatial pattern in Salmonella seropositivity in Danish farms, this may suggest an important influence of common contaminated pig supply or the effect of a farm adviser in that specific region.
In contrast to previous studies where there was a correlation between the size of the farm and Salmonella status [Reference Correia-Gomes16], no statistically significant association was found in the present study, although the proportion of positive farms tended to be higher in farms with >1000 sows.
The main serovars identified in this study differed from previous reports in the country [Reference Parada6, Reference Ibar17] and in the region [Reference Kich18], and from those found in a European survey [2]. All those studies reported the isolation of a large number of serovars, in contrast to our results in which S. Typhimurium and S. Derby where the most prevalent serotypes. One possible explanation is the sampling methodology, since most of the studies described previously were focused on slaughterhouse-based sampling and the approach used in the present study may represent better the in-farm pig situation, since it prevents the contamination that has frequently been reported when samples are collected at the abattoir [Reference De Busser1], or even on the farm using environmental samples [Reference Kich18].
The main serovars isolated, S. Typhimurium and S. Derby, are in agreement with reports in North America and European countries [Reference Sanchez3] and partly with Brazil [Reference Kich18], but differ from reports from Chile where Villamil et al. [Reference Villamil, González and Ruiz5] showed S. Infantis as more prevalent, a serotype that was scarcely found in the present study, and by Kich et al. [Reference Kich18]. It also highlights the potential role of pigs as reservoirs of infection for the general public, since S. Typhimurium has been reported as the main serotype in human infections in Latin America [Reference Campos7].
Consistent with previous studies [Reference Benschop8, Reference Correia-Gomes16], one of the most significant explicative variables for the herd Salmonella status in the regression model was nutrition. This could be due not only to the potential direct effect of feed as a carrier of Salmonella, but also due to the risks associated with the logistics of feed delivery [Reference Davies and Wales19], especially in nearby farms. Not unexpectedly, the presence of diarrhoea in fattening pigs was also associated with increased odds of testing positive for Salmonella. However, this result must be considered carefully, because there are several aetiologies that can be responsible for this sign that were not studied. Moreover, there were farms where Salmonella was isolated but diarrhoea was not reported that could be due to the existence of subclinical infections as previously suggested [Reference Guenther20].
A controversial point is the use of footbaths as a measure for Salmonella control in farms. While some studies highlight the usefulness of this measure [Reference Hautekiet10], others proposed that it may contribute to the spread of the pathogen [Reference Aury21]. In our study, there was no statistical association between the presence of Salmonella and the use of footbaths in fattening facilities, which suggests that its use per se should not be considered a protective factor. However, other factors associated with proper footbath management in addition to its mere use (renewal, concentration and activity of the active agent) may be more important to decrease the risk of Salmonella infection.
To the best of our knowledge, this is the first study of herd prevalence and spatial epidemiology of Salmonella in pigs in Argentina. The methodology described in the present study allowed detection of the presence of important Salmonella serovars in slaughter-age pigs that have often been considered of public health concern, the distribution of these serovars, as well as a spatial clustering of Salmonella-positive herds in a concentrated pig production region. Even though a reduced sample size was analysed, what could compromise the power of the analysis to detect certain associations, and considering the possible biases introduced by the use of non-probabilistic method, our results highlight the need to evaluate the microbiological controls at the farm level, and demonstrates the usefulness of the spatial tools to identify areas of greatest risk when processing pork at slaughterhouse, which could contribute to increased food-safety of pork products. However, future research is needed to evaluate the stability in time of this spatial distribution.
ACKNOWLEDGEMENTS
The authors thank Dr M. I. Caffer and Dr M. Moroni for their assistance in strain identification and subtyping, and the Antigens and Antisera Service, INEI- ANLIS ‘Carlos G. Malbrán’, Buenos Aires, Argentina. In addition, we thank M. Monterubbianesi and M. Herrera from SENASA for providing farm access and sampling assistance, and especially to P. Camacho and Dr A. Alba for their technical collaboration.
DECLARATION OF INTEREST
None.







