I. INTRODUCTION
Oxfendazole (sold under the brand name Synanthic®) is a broad spectrum benzimidazole anthelmintic. Its main use is for protecting livestock against roundworm, strongyles (bloodworms), and pinworms. The systematic name (CAS Registry Number 53716-50-0) is methyl N-[6-(benzenesulfinyl)-1H-benzimidazol-2-yl]carbamate. A two-dimensional (2D) molecular diagram is shown in Figure 1.

Figure 1. The 2D molecular structure of oxfendazole.
Suspensions of oxfendazole nanocrystals have been studied for use as an oral formulation, and an X-ray powder diffraction pattern of the API has been provided (Sun et al., Reference Sun, Chen, Zhao, Zhou, Zhang, Wang and Xie2022).
This work was carried out as part of a project (Kaduk et al., Reference Kaduk, Crowder, Zhong, Fawcett and Suchomel2014) to determine the crystal structures of large-volume commercial pharmaceuticals, and include high-quality powder diffraction data for them in the Powder Diffraction File (Gates-Rector and Blanton, Reference Gates-Rector and Blanton2019).
II. EXPERIMENTAL
Oxfendazole was a commercial reagent, purchased from TargetMol (Lot #113808), and was used as-received. The white powder was packed into a 1.5-mm diameter Kapton capillary, and rotated during the measurement at ~50 Hz. The powder pattern was measured at 295 K at beamline 11-BM (Antao et al., Reference Antao, Hassan, Wang, Lee and Toby2008; Lee et al., Reference Lee, Shu, Ramanathan, Preissner, Wang, Beno, Von Dreele, Ribaud, Kurtz, Antao, Jiao and Toby2008; Wang et al., Reference Wang, Toby, Lee, Ribaud, Antao, Kurtz, Ramanathan, Von Dreele and Beno2008) of the Advanced Photon Source at Argonne National Laboratory using a wavelength of 0.458208(2) Å from 0.5° to 50° 2θ with a step size of 0.001° and a counting time of 0.1 s/step. The high-resolution powder diffraction data were collected using twelve silicon crystal analyzers that allow for high angular resolution, high precision, and accurate peak positions. A silicon (NIST SRM 640c) and alumina (SRM 676a) standard (ratio Al2O3:Si = 2:1 by weight) was used to calibrate the instrument and refine the monochromatic wavelength used in the experiment.
The pattern was indexed using both JADE Pro (MDI, 2022) and N-TREOR (Altomare et al., Reference Altomare, Cuocci, Giacovazzo, Moliterni, Rizzi, Corriero and Falcicchio2013) on a primitive monoclinic unit cell with a = 18.85890, b = 10.40423, c = 7.25538 Å, β = 91.553°, V = 1423.1 Å3, and Z = 4. A reduced cell search in the Cambridge Structural Database (Groom et al., Reference Groom, Bruno, Lightfoot and Ward2016) yielded 13 hits, but no structures of oxfendazole derivatives. The suggested space group was P21/c, which was confirmed by successful solution and refinement of the structure.
An oxfendazole molecule was downloaded from PubChem (Kim et al., Reference Kim, Chen, Cheng, Gindulyte, He, He, Li, Shoemaker, Thiessen, Yu, Zaslavsky, Zhang and Bolton2019) as Conformer3D_CID_40854.sdf. It was converted to a *.mol2 file using Mercury (Macrae et al., Reference Macrae, Sovago, Cottrell, Galek, McCabe, Pidcock, Platings, Shields, Stevens, Towler and Wood2020). The structure was solved by Monte Carlo simulated annealing techniques as implemented in EXPO2014 (Altomare et al., Reference Altomare, Cuocci, Giacovazzo, Moliterni, Rizzi, Corriero and Falcicchio2013).
Rietveld refinement was carried out using GSAS-II (Toby and Von Dreele, Reference Toby and Von Dreele2013). Only the 1.0–25.0° portion of the pattern was included in the refinement (d min = 1.058 Å). The region 1.53–2.07° 2θ, which contained a broad peak from the Kapton capillary, was excluded. All non-H bond distances and angles were subjected to restraints, based on a Mercury/Mogul Geometry check (Bruno et al., Reference Bruno, Cole, Kessler, Luo, Motherwell, Purkis, Smith, Taylor, Cooper, Harris and Orpen2004; Sykes et al., Reference Sykes, McCabe, Allen, Battle, Bruno and Wood2011). The Mogul average and standard deviation for each quantity were used as the restraint parameters. The restraints contributed 3.1% to the final χ 2. The hydrogen atoms were included in calculated positions, which were recalculated during the refinement using Materials Studio (Dassault, 2021). The U iso of the heavy atoms were grouped by chemical similarity. The U iso for the H atoms were fixed at 1.2× the U iso of the heavy atoms to which they are attached. The peak profiles were described using the generalized microstrain model. The background was modeled using a 6-term shifted Chebyshev polynomial, and a peak at 5.87° 2θ to model the scattering from the Kapton capillary and any amorphous component.
The final refinement of 94 variables using 23,501 observations and 57 restraints yielded the residuals R wp = 0.0785 and GOF = 1.56. The largest peak (1.49 Å from C18) and hole (1.36 Å from C19) in the difference Fourier map were 0.23(5) and −0.21(5) eÅ−3, respectively. The largest errors in the difference plot (Figure 2) are in the shape of the strong lowest-angle (1 0 0) peak.
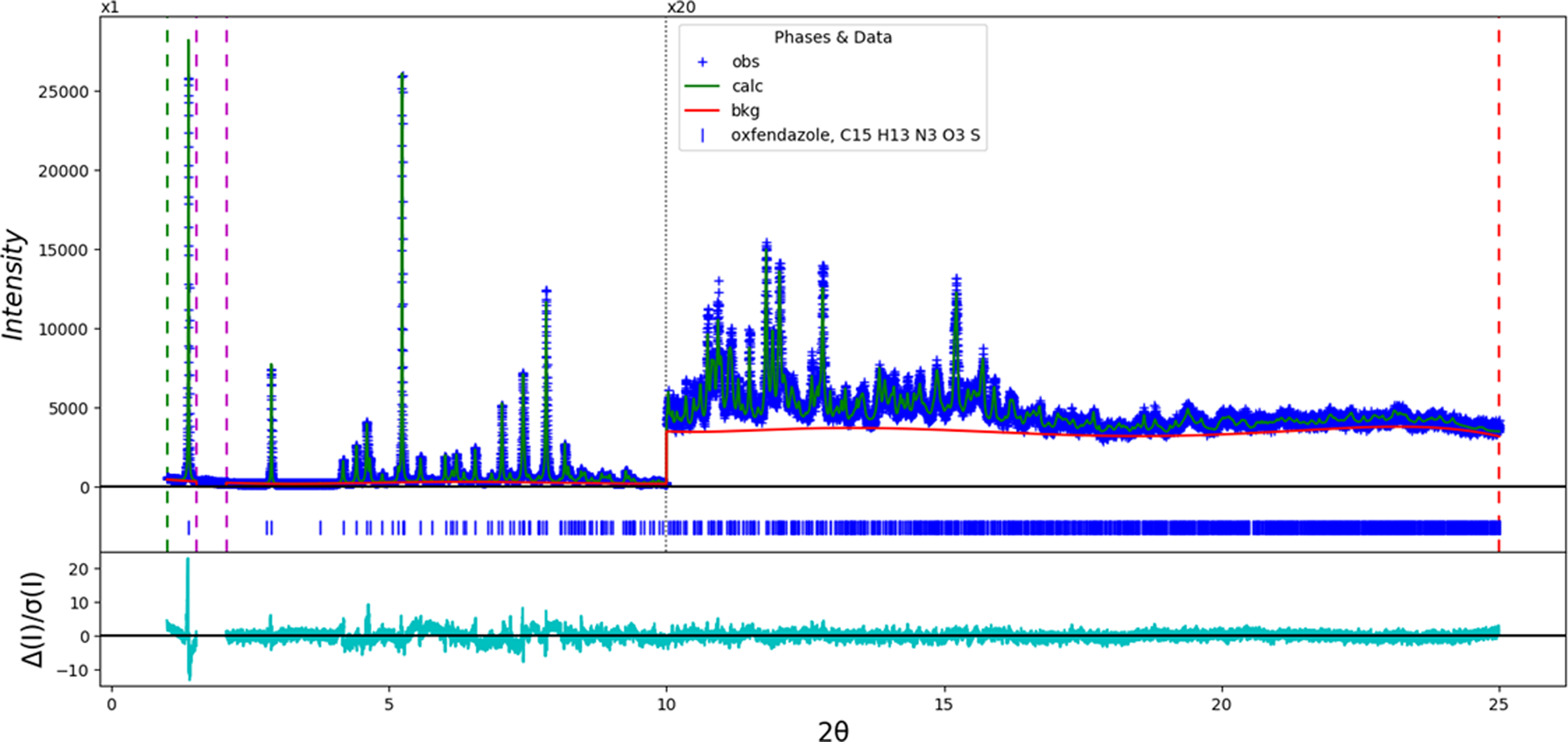
Figure 2. The Rietveld plot for the refinement of oxfendazole. The blue crosses represent the observed data points, and the green line is the calculated pattern. The cyan curve is the normalized error plot. The red curve indicates the background. The vertical scale has been multiplied by a factor of 20× for 2θ > 10.0°. The row of blue tick marks indicates the calculated reflection positions.
The crystal structure was optimized using VASP (Kresse and Furthmüller, Reference Kresse and Furthmüller1996) (fixed experimental unit cell) through the MedeA graphical interface (Materials Design, 2016). The calculation was carried out on 16 2.4 GHz processors (each with 4 GB RAM) of a 64-processor HP Proliant DL580 Generation 7 Linux cluster at North Central College. The calculation used the GGA-PBE functional, a plane wave cutoff energy of 400.0 eV, and a k-point spacing of 0.5 Å−1 leading to a 1 × 2 × 2 mesh, and took ~15.1 h. A single-point density functional theory calculation (fixed experimental cell) and population analysis were carried out using CRYSTAL17 (Dovesi et al., Reference Dovesi, Erba, Orlando, Zicovich-Wilson, Civalleri, Maschio, Rerat, Casassa, Baima, Salustro and Kirtman2018). The basis sets for the H, C, N, and O atoms in the calculation were those of Gatti et al. (Reference Gatti, Saunders and Roetti1994), and that for S was that of Peintinger et al. (Reference Peintinger, Vilela Oliveira and Bredow2013). The calculations were run on a 3.5 GHz PC using 8 k-points and the B3LYP functional, and took ~1.9 h.
III. RESULTS AND DISCUSSION
The synchrotron powder pattern of this study matches the pattern for oxfendazole (in a single-column figure) reported by Sun et al. (Reference Sun, Chen, Zhao, Zhou, Zhang, Wang and Xie2022) well enough to conclude that they represent the same material (Figure 3), and thus that this material is a representative sample. Sun et al. did not start at an angle low-enough to measure the lowest-angle peak. The root-mean-square Cartesian displacement between the Rietveld-refined and DFT-optimized structures is 0.183 Å (Figure 4), and the maximum difference is 0.426 Å, at O4. The excellent agreement is strong evidence that the structure is correct (van de Streek and Neumann, Reference van de Streek and Neumann2014). This discussion concentrates on the DFT-optimized structure. The asymmetric unit (with atom numbering) is illustrated in Figure 5. The best view of the crystal structure is down the b-axis (Figure 6). The crystal structure consists of stacks of the planar portions of the L-shaped molecules. The mean plane of the benzimidazole ring system is approximately 8,1,10, and the plane of the phenyl ring is approximately 13,8,−5. The Mercury Aromatics Analyser indicates three moderate interactions: one π–π with a distance of 3.80 Å, one H–π with a distance of 5.32 Å, and one side-side with a distance of 6.42 Å.

Figure 3. Comparison of the synchrotron pattern of oxfendazole (black) to that reported by Sun et al. (Reference Sun, Chen, Zhao, Zhou, Zhang, Wang and Xie2022; green). The literature pattern, measured using CuKα radiation, was digitized using UN-SCAN-IT (Silk Scientific, 2013), and converted to the synchrotron wavelength of 0.458208 Å using JADE Pro (MDI, 2022), which was also used to generate the image.

Figure 4. Comparison of the Rietveld-refined (red) and VASP-optimized (blue) structures of oxfendazole. The rms Cartesian displacement is 0.183 Å. Image generated using Mercury (Macrae et al., Reference Macrae, Sovago, Cottrell, Galek, McCabe, Pidcock, Platings, Shields, Stevens, Towler and Wood2020).

Figure 5. The asymmetric unit of oxfendazole, with the atom numbering. The atoms are represented by 50% probability spheroids/ellipsoids. Image generated using Mercury (Macrae et al., Reference Macrae, Sovago, Cottrell, Galek, McCabe, Pidcock, Platings, Shields, Stevens, Towler and Wood2020).

Figure 6. The crystal structure of oxfendazole, viewed down the b-axis. Image generated using Diamond (Crystal Impact, 2022).
All of the bond distances and bond angles fall within the normal ranges indicated by a Mercury/Mogul Geometry check (Macrae et al., Reference Macrae, Sovago, Cottrell, Galek, McCabe, Pidcock, Platings, Shields, Stevens, Towler and Wood2020). The C8–C11–C10 angle of 115.8° in the benzimidazole ring system is flagged as slightly unusual (average = 120.1(14)°, Z-score = 3.1). The torsion angles involving rotation about the C17–N7 bond are flagged as unusual. These lie in a minor population of a distribution with the majority rotated by 180°. They represent the orientation of the side chain with respect to the benzimidazole ring system, and indicate that the conformation of the molecule in the solid state is unusual.
Quantum chemical geometry optimization of the oxfendazole molecule (DFT/B3LYP/6-31G*/water) using Spartan ‘18 (Wavefunction, 2020) indicated that in the solid state it is in essentially a local minimum energy conformation (rms displacement = 0.174 Å). A conformational analysis (MMFF force field) indicates that the global minimum-energy conformation has the side chain rotation approximately 180° from the observed conformation. Intermolecular interactions seem to be important in determining the solid-state conformation.
Analysis of the contributions to the total crystal energy of the structure using the Forcite module of Materials Studio (Dassault, 2021) suggests that the intramolecular deformation energy is dominated by angle distortion terms, as might be expected for a fused ring system. The intermolecular energy is dominated by electrostatic attractions, which in this force field analysis include hydrogen bonds. The hydrogen bonds are better analyzed using the results of the DFT calculation.
Only weak hydrogen bonds are present in the structure (Table I). There are two N–H⋯O hydrogen bonds to the carbonyl group O4. The energies of these bonds were calculated using the correlation of Wheatley and Kaduk (Reference Wheatley and Kaduk2019). N7–H27 makes an additional intermolecular N–H⋯N hydrogen bond to N5. Several C–H⋯O hydrogen bonds (one intramolecular) also contribute to the lattice energy.
TABLE I. Hydrogen bonds (CRYSTAL17) in oxfendazole.

a Intramolecular.
The volume enclosed by the Hirshfeld surface of oxfendazole (Figure 7, Hirshfeld, Reference Hirshfeld1977; Turner et al., Reference Turner, McKinnon, Wolff, Grimwood, Spackman, Jayatilaka and Spackman2017) is 348.53 Å3, 97.96% of 1/4 the unit cell volume. The packing density is thus fairly typical. The only significant close contacts (red in Figure 7) involve the hydrogen bonds. The volume/non-hydrogen atom is smaller than normal at 16.2 Å3.

Figure 7. The Hirshfeld surface of oxfendazole. Intermolecular contacts longer than the sums of the van der Waals radii are colored blue, and contacts shorter than the sums of the radii are colored red. Contacts equal to the sums of radii are white. Image generated using CrystalExplorer (Turner et al., Reference Turner, McKinnon, Wolff, Grimwood, Spackman, Jayatilaka and Spackman2017).
The Bravais–Friedel–Donnay–Harker (Bravais, Reference Bravais1866; Friedel, Reference Friedel1907; Donnay and Harker, Reference Donnay and Harker1937) morphology suggests that we might expect platy morphology for oxfendazole, with {100} as principal faces. A second-order spherical harmonic preferred orientation model was included in the refinement. The texture index was 1.000(0), indicating that preferred orientation was not significant for this rotated capillary specimen. The powder pattern of oxfendazole from this synchrotron data set has been submitted to ICDD for inclusion in the Powder Diffraction File.
IV. DEPOSITED DATA
The Crystallographic Information Framework (CIF) files containing the results of the Rietveld refinement (including the raw data) and the DFT geometry optimization were deposited with the ICDD. The data can be requested at [email protected].
ACKNOWLEDGMENTS
The use of the Advanced Photon Source at Argonne National Laboratory was supported by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract No. DE-AC02-06CH11357. This work was partially supported by the International Centre for Diffraction Data. We thank Lynn Ribaud and Saul Lapidus for their assistance in the data collection.
CONFLICTS OF INTEREST
The authors have no conflicts of interest to declare.











