Published online by Cambridge University Press: 01 January 2025
Cognitive diagnostic models (CDMs) are discrete latent variable models popular in educational and psychological measurement. In this work, motivated by the advantages of deep generative modeling and by identifiability considerations, we propose a new family of DeepCDMs, to hunt for deep discrete diagnostic information. The new class of models enjoys nice properties of identifiability, parsimony, and interpretability. Mathematically, DeepCDMs are entirely identifiable, including even fully exploratory settings and allowing to uniquely identify the parameters and discrete loading structures (the “Q\documentclass[12pt]{minimal}\usepackage{amsmath}\usepackage{wasysym}\usepackage{amsfonts}\usepackage{amssymb}\usepackage{amsbsy}\usepackage{mathrsfs}\usepackage{upgreek}\setlength{\oddsidemargin}{-69pt}\begin{document}$$\textbf{Q}$$\end{document}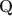 -matrices”) at all different depths in the generative model. Statistically, DeepCDMs are parsimonious, because they can use a relatively small number of parameters to expressively model data thanks to the depth. Practically, DeepCDMs are interpretable, because the shrinking-ladder-shaped deep architecture can capture cognitive concepts and provide multi-granularity skill diagnoses from coarse to fine grained and from high level to detailed. For identifiability, we establish transparent identifiability conditions for various DeepCDMs. Our conditions impose intuitive constraints on the structures of the multiple Q\documentclass[12pt]{minimal}\usepackage{amsmath}\usepackage{wasysym}\usepackage{amsfonts}\usepackage{amssymb}\usepackage{amsbsy}\usepackage{mathrsfs}\usepackage{upgreek}\setlength{\oddsidemargin}{-69pt}\begin{document}$$\textbf{Q}$$\end{document}
-matrices”) at all different depths in the generative model. Statistically, DeepCDMs are parsimonious, because they can use a relatively small number of parameters to expressively model data thanks to the depth. Practically, DeepCDMs are interpretable, because the shrinking-ladder-shaped deep architecture can capture cognitive concepts and provide multi-granularity skill diagnoses from coarse to fine grained and from high level to detailed. For identifiability, we establish transparent identifiability conditions for various DeepCDMs. Our conditions impose intuitive constraints on the structures of the multiple Q\documentclass[12pt]{minimal}\usepackage{amsmath}\usepackage{wasysym}\usepackage{amsfonts}\usepackage{amssymb}\usepackage{amsbsy}\usepackage{mathrsfs}\usepackage{upgreek}\setlength{\oddsidemargin}{-69pt}\begin{document}$$\textbf{Q}$$\end{document}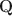 -matrices and inspire a generative graph with increasingly smaller latent layers when going deeper. For estimation and computation, we focus on the confirmatory setting with known Q\documentclass[12pt]{minimal}\usepackage{amsmath}\usepackage{wasysym}\usepackage{amsfonts}\usepackage{amssymb}\usepackage{amsbsy}\usepackage{mathrsfs}\usepackage{upgreek}\setlength{\oddsidemargin}{-69pt}\begin{document}$$\textbf{Q}$$\end{document}
-matrices and inspire a generative graph with increasingly smaller latent layers when going deeper. For estimation and computation, we focus on the confirmatory setting with known Q\documentclass[12pt]{minimal}\usepackage{amsmath}\usepackage{wasysym}\usepackage{amsfonts}\usepackage{amssymb}\usepackage{amsbsy}\usepackage{mathrsfs}\usepackage{upgreek}\setlength{\oddsidemargin}{-69pt}\begin{document}$$\textbf{Q}$$\end{document}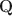 -matrices and develop Bayesian formulations and efficient Gibbs sampling algorithms. Simulation studies and an application to the TIMSS 2019 math assessment data demonstrate the usefulness of the proposed methodology.
-matrices and develop Bayesian formulations and efficient Gibbs sampling algorithms. Simulation studies and an application to the TIMSS 2019 math assessment data demonstrate the usefulness of the proposed methodology.
To send this article to your Kindle, first ensure no-reply@cambridge.org is added to your Approved Personal Document E-mail List under your Personal Document Settings on the Manage Your Content and Devices page of your Amazon account. Then enter the ‘name’ part of your Kindle email address below. Find out more about sending to your Kindle. Find out more about saving to your Kindle.
Note you can select to save to either the @free.kindle.com or @kindle.com variations. ‘@free.kindle.com’ emails are free but can only be saved to your device when it is connected to wi-fi. ‘@kindle.com’ emails can be delivered even when you are not connected to wi-fi, but note that service fees apply.
Find out more about the Kindle Personal Document Service.
To save this article to your Dropbox account, please select one or more formats and confirm that you agree to abide by our usage policies. If this is the first time you used this feature, you will be asked to authorise Cambridge Core to connect with your Dropbox account. Find out more about saving content to Dropbox.
To save this article to your Google Drive account, please select one or more formats and confirm that you agree to abide by our usage policies. If this is the first time you used this feature, you will be asked to authorise Cambridge Core to connect with your Google Drive account. Find out more about saving content to Google Drive.