To the Editor—New Delhi metallo-β-lactamase (NDM) is one of the main globally described carbapenemases. It was first reported in 2009 in India.Reference Yong, Toleman, Giske, Cho, Sundman, Lee and Walsh1 Providencia rettgeri was first reported in Brazil in 2013.Reference Carvalho-Assef, Pereira and Albano2 NDM emergence has been described in Brazil among gram-negative bacteria related to infection or the environment.Reference Rozales, Magagnin and Campos3–Reference Campana, Montezzi, Paschoal and Picão5 Here, we describe an outbreak of NDM-1–producing Klebsiella pneumoniae (KPN) strains ST340 and ST2570 in 50 single isolates from 2 Brazilian hospitals between May 2017 and July 2018 (Figure 1). These hospitals belong to a regional medical complex with different locations and professionals. Their microbiology laboratory has applied the same metallo-β-lactamase (MβL) screening procedure with commercial carbapenem disks with and without EDTA (0.1 M) since 2016. Patients admitted are routinely screened for the presence of multidrug-resistant Enterobacteriaceae. The lab identified 50 KPN-MβL–positive carbapenem-resistant single strains using the Vitek MS system (bioMeriéux, Marcy-I’Étoile, France). One sample per patient was considered, and the bla NDM-1 gene was detected using multiplex real-time polymerase chain reaction (qPCR).Reference Monteiro, Widen, Pignatari, Kubasek and Silbert6
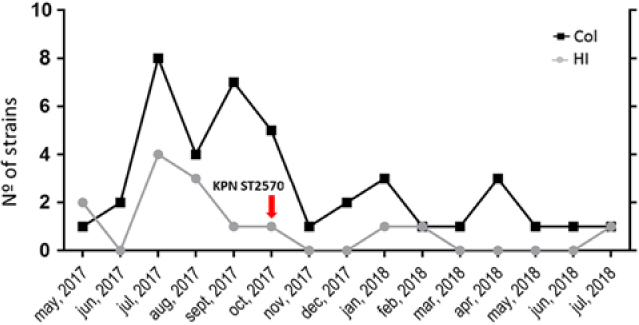
Fig. 1. New Delhi metallo-β-lactamase producing Klebsiella pneumoniae (NDM KPN) case distribution along time.
Overall, 45 isolates were identified from hospital 1, and 5 were identified from hospital 2. Among them, 41 were from surveillance cultures (rectal swabs) and 9 were from clinical samples (5 urine, 3 blood and 1 tracheobronchial aspirate). At the moment of isolation, 14 of these 50 patients had a diagnosis of hospital-acquired infection (HAI) according to established criteria (7 nonventilation pneumonia, 4 bloodstream, 2 catheter, and 1 skin and soft-tissue infections). In 5 of 14 patients, NDM-KPN was potentially associated with the HAI (3 blood and 2 urine cultures). In 5 of the 14 HAI patients, other microorganisms were considered causative agents, and in 4 these patients, no agents were isolated from infections (NDM KPN from surveillance only). At admission, 7 of the 14 HAI patients presented with community infections, 4 with orthopedic or trauma, and 2 with neurological conditions. Also, 5 of these 14 HAI patients had multiple comorbidities. Notably, 7 of these 14 patients were later discharged with improved conditions, and 7 of these patients died during their hospital stay.
Antimicrobial susceptibility testing was performed by Vitek 2 System (bioMeriéux), and high-level resistance was detected for all β-lactams, ciprofloxacin, and gentamicin (ie, using the guidelines of the Clinical Laboratory Standards Institute [CLSI]). All strains were susceptible to colistin (minimum inhibitory concentration [MIC], ≤2 µg/mL), and 40 of 50 were resistant to tigecycline (MIC, >2 µg/mL) (ie, using the European Committee on Antimicrobial Susceptibility Testing (EUCAST) criteria). ESβLs (ie, bla TEM, bla SHV, and bla CTX-M) and quinolone resistance genes (qnrS) were detected in all isolates. A molecular investigation was performed using pulsed field gel-electrophoresis (PFGE), and multilocus sequence typing (MLST). PFGE showed 2 different patterns, A and B. Pattern A was found in 44 of 45 strains from hospital 1 and in 5 of 5 strains from hospital 2. Only 1 strain from hospital 1 showed a distinct PFGE pattern (pattern B). MLST was performed in 5 KPN strains, 4 belonging to pattern A and 1 belonging to pattern B. Also, 4 pattern A strains belonged to the same ST340, clonal complex 258 (CC258). The pattern B strain belonged to ST2570, a different and nonphylogenetically related strain.
The first (or index) case occurred in hospital 1 in May 2017 on a female patient hospitalized at an isolation unit. This patient had many comorbidities (ie, diabetes, hypertension, Alzheimer disease, and previous hemorrhagic stroke) and was admitted to the hospital due to urinary tract infection. On May 21, 2017, she was diagnosed with nonventilation pneumonia and presented both KPN and Enterobacter cloacae from blood cultures. Despite therapy, she died during hospitalization on July 1, 2017. The KPN isolate belonged to PFGE pattern A and ST340.
Various studies have demonstrated that the KPN phylogenetic lineage belonging to CC258 (including ST258, ST11, ST340, and ST437) is vastly adapted to human populations and to hospital infections.Reference Castanheira, Costello, Deshpande and Jones7 Previous reports of KPN ST340 and ST11 carrying NDM and other resistance mechanisms (ESβLs and qnr genes) have been documented in Brazil.Reference Aires, Pereira and de Araujo4, Reference Campana, Montezzi, Paschoal and Picão5 The emergence and spread of NDM KPN is worrying and represents a new worldwide challenge because it may carry several high-level antimicrobial resistance mechanisms.
The KPN pattern B and ST2570 strain was recovered from a blood culture of a patient hospitalized on a different unit in hospital 1 on October 6, 2017. This patient was initially hospitalized on September 20, 2017, due to a skin and soft-tissue infection and various comorbidities (ie, human immunodeficiency virus [HIV], treated tuberculosis, and chronic obstructive pulmonary disease [COPD]) and with many previous hospital passages. The blood culture was positive for NDM KPN on October 6, 2017, and the HAI was a skin and soft-tissue infection. Unfortunately, this patient was sent to the intensive care unit (ICU) and died 15 days later, despite medical care and antimicrobial therapy (including meropenem and polymixin B). Notably, the detection of the KPN ST2570 isolate seems relevant. Eibach et alReference Eibach, Dekker and Gyau Boahen8 described ESβL-producing KPN ST2570 in samples from local and imported poultry in Ghana. To the best of our knowledge, this is the first human case (bloodstream infection and sepsis) caused by an NDM-KPN ST2570. It has been shown that NDM plasmid-bearing microorganisms might have significant environmental presence,Reference Nordmann, Poirel, Walsh and Livermore9, Reference Walsh, Weeks, Livermore and Toleman10 which could be gene sources to other species. We do not know which role the ST2570 isolate played in this outbreak, but it presence raises an interesting discussion, especially in a patient with multiple comorbidities and admittances. Could distinct KPN lineages adapted to poultry be the linkage between CC258 and environmental NDM isolates?
We report the first major outbreak of NDM KPN in hospitals from a single city in Brazil. Most isolates belonged to a well-adapted and documented CC258 (ST340). One case was caused by a distinct and not phylogenetically related ST2570. This outbreak highlights the relevance of monoclonal isolate dissemination between environments, despite hospital control policies. Apparently, monoclonal dissemination of well-adapted isolates takes place before plasmid dissemination to other clones or species.
Author ORCIDs
Jussimara Monteiro 0000-0002-1230-4816
Financial support
No financial support was provided relevant to this article.
Conflicts of interest
All authors report no conflicts of interest relevant to this article.



