Introduction
Enterotoxigenic Escherichia coli (ETEC) is a diarrhoeagenic E. coli that causes diarrhoea not only in humans but also in various animals, such as pigs, cattle and sheep [Reference Dubreuil, Isaacson and Schifferli1]. Regardless of host, colonisation of the intestinal epithelia is an essential first step in the pathogenesis of ETEC infection mediated by a variety of colonisation factors (CFs) [Reference Vipin Madhavan and Sakellaris2]. Subsequently, ETEC secretes host-damaging toxins such as heat-labile (LT) and/or heat-stable (ST) enterotoxins, which give rise to intestinal symptoms such as diarrhoea [Reference Mirhoseini, Amani and Nazarian3, Reference Sinha4].
ST-producing ETEC O169:H41 (O169) was first identified as an aetiological serotype in human foodborne cases in Japan [Reference Nishikawa5]. Previously, we reported that the O169 strain YN10 harbours the unstable large plasmid pEntYN10, which codes for genes of three CFs, CS6 and two novel CFs resembling CS8 (so-called CFA/III) and F4 (so-called K88). The operon coding the novel K88-like CF (hereinafter, K88O169) has two paralogous major adhesin-like subunits, faeG1 and faeG2, which have 37–44% amino acid homology with faeG of the original K88 [Reference Ban6].
K88 and its related CFs have been found with bacteria isolated from a variety of hosts. K88 was initially identified as a CF of swine ETEC [Reference Ørskov7, Reference Johnson and Nolan8], whereas CS31A was reported as a K88-related CF from bovine ETEC [Reference Girardeau9]. Thus, E. coli strains with analogous CFs were isolated from multiple hosts, suggesting that K88-related CFs can lead to colonisation in various host species. Furthermore, an ETEC/Shiga toxin-producing E. coli (STEC) hybrid strain was recently isolated from a patient with hemolytic-uremic syndrome (HUS); the organism possessed an F4-like adhesin of the protein sequence that was similar to CS31A of bovine ETEC [Reference Michelacci10]. The presence of multiple CFs in O169 may indicate the potential for multi-host transmission: a complex zoonotic network may be supported, and this contradicts the one-to-one host-pathogen relationship that has been commonly accepted for ETEC.
This study examined the seroepidemiological status of livestock against K88O169 as a preliminary investigation to test this hypothesis. Antibody titres against K88O169 of pigs and cattle reared in various areas in Japan were examined by an indirect ELISA method using recombinant proteins of the adhesins, FaeG.
Materials and methods
Sample collection
In this study, sera of 200 pigs and 105 cattle collected from June 2019 to March 2020 were kindly provided by the Osaka City Meat Hygiene Inspection Center for serological assays. This facility is partnered with the slaughterhouse and is responsible for hygiene inspections of meat supplied to the metropolitan area and also conducts research on livestock animals transported from a wide range of regions in Japan. Breeders and the prefectures where the animals were raised are listed in Supporting Tables S1 and S2.
Recombinant proteins
Recombinant proteins were created using the pET expression system (Novagen). The codon-optimised typical faeG gene of the original K88 (K88ac variant, accession ID: AJ616239) [Reference Verdonck11] was synthesised by Eurofins Genomics KK (Tokyo, Japan). faeG1 and faeG2 genes of K88O169 were amplified by PCR with primers for the O169 plasmid pEntYN10 [Reference Ban6] as shown in Supporting Table S3. The primers were designed to remove signal peptides predicted using SignalP 5.0 web-based resource [Reference Almagro Armenteros12] from the recombinant proteins. These gene products and pET30a vector (Novagen) were digested with SalI and NotI restriction enzymes and ligated. The resulting plasmids (pET30a-faeG, pET30a-faeG1 and pET30a-faeG2) were recovered by transformation into E. coli BL21 (DE3) with selection for kanamycin resistance. After the bacteria were grown at 37 °C for 3 h, protein expression was induced with 1 mm of isopropyl-β-D-thiogalactopyranoside (IPTG; Takara Bio) at the final concentration at 25 °C. After overnight incubation, the cells were solubilised with 8 M urea. The recombinant protein was purified using the His60 Ni gravity column purification kit (Takara Bio) according to the manual instruction. Finally, the solution was exchanged with phosphate-buffered saline (PBS) using Amicon Ultra-15 10 K Centrifugal Filter Devices (Merck). Purity was confirmed by the absence of contamination bands on SDS-PAGE.
Antiserum
Antiserum to the original K88 was purchased from Denka Seiken (E. coli K88 rabbit antiserum). Rabbit polyclonal antibodies against FaeG1 and FaeG2 developed by Eurofins Genomics Antibody Service were used as positive controls.
Indirect ELISA
Sample sera were examined for the antibodies against FaeG of K88 and FaeG1 and FaeG2 of K88O169 with indirect ELISA methods. Purified protein was coated on 96-well half-area plates (Corning) at 0.1 μg/ml with coating buffer (13 mm Na2CO3, 35 mm NaHCO3) overnight at 4 °C. After washing three times with PBS-T (PBS containing 0.05% Tween-20), plates were blocked by adding blocking buffer (0.4% Block Ace, Dainippon Pharmaceutical) and incubated for 1–2 h. After three washes, sample sera (1:200 dilution in PBS-T) and antibodies (1:1000 dilution) as a positive control were added to the plate and incubated for 1 h. After three washes, a 1: 30 000 dilution of the secondary antibody solution was added to each well and incubated for 1 h. The secondary antibodies used for ELISA were as follows: HRP-linked anti-whole rabbit IgG donkey serum (GE Healthcare, #NA934) for detection of the positive control, HRP-linked anti-pig whole IgG goat antibodies (Abcam, #ab6915) for porcine samples, and HRP-linked anti-cow whole IgG goat antibodies (Abcam, #ab102154) for bovine samples. After three washes, TMB substrate (TMB Microwell Peroxidase Substrate System, KPL) was added to the plates, incubated for 5 min in the dark, stopped with 1 M H2SO4, and measured at wavelengths of 450 nm/550 nm with a microplate reader (Wallac 1420 ARVOsx, Perkin Elmer). Sample to positive (S/P) ratio (%) was calculated as follows: (Sample serum – blank)/(Positive control antibody – blank). S/P ratio >0.4 was considered as positive [Reference Yahara13, Reference Li14].
Statistical analysis
The results were statistically analysed using Prism software (GraphPad, ver. 8.4.3). Fisher's exact test was employed to assess the association between porcine and bovine seroprevalence.
Results
Anti-FaeG1 and -FaeG2 antibodies in porcine and bovine sera were measured to determine whether K88O169 may be another CF for infection of pigs or cattle. Among the samples tested, a portion of the porcine (18.0%, 36/200) and bovine (17.1%, 18/105) sera reacted to one or both of FaeG1 and FaeG2 antigens of K88O169 (Table 1, Fig. 1). The number of the 54 positive sera samples reacting to FaeG1, FaeG2, and both antigens was 29 (54%), 9 (17%) and 16 (29%), respectively. As 45 and 25 sera reacted with FaeG1 and FaeG2, respectively, anti-FaeG1 was significantly more prevalent than anti-FaeG2 (P < 0.0001). These results were not biased by collection location as sera samples were collected from various regions in Japan (Supporting Tables S1 and S2).
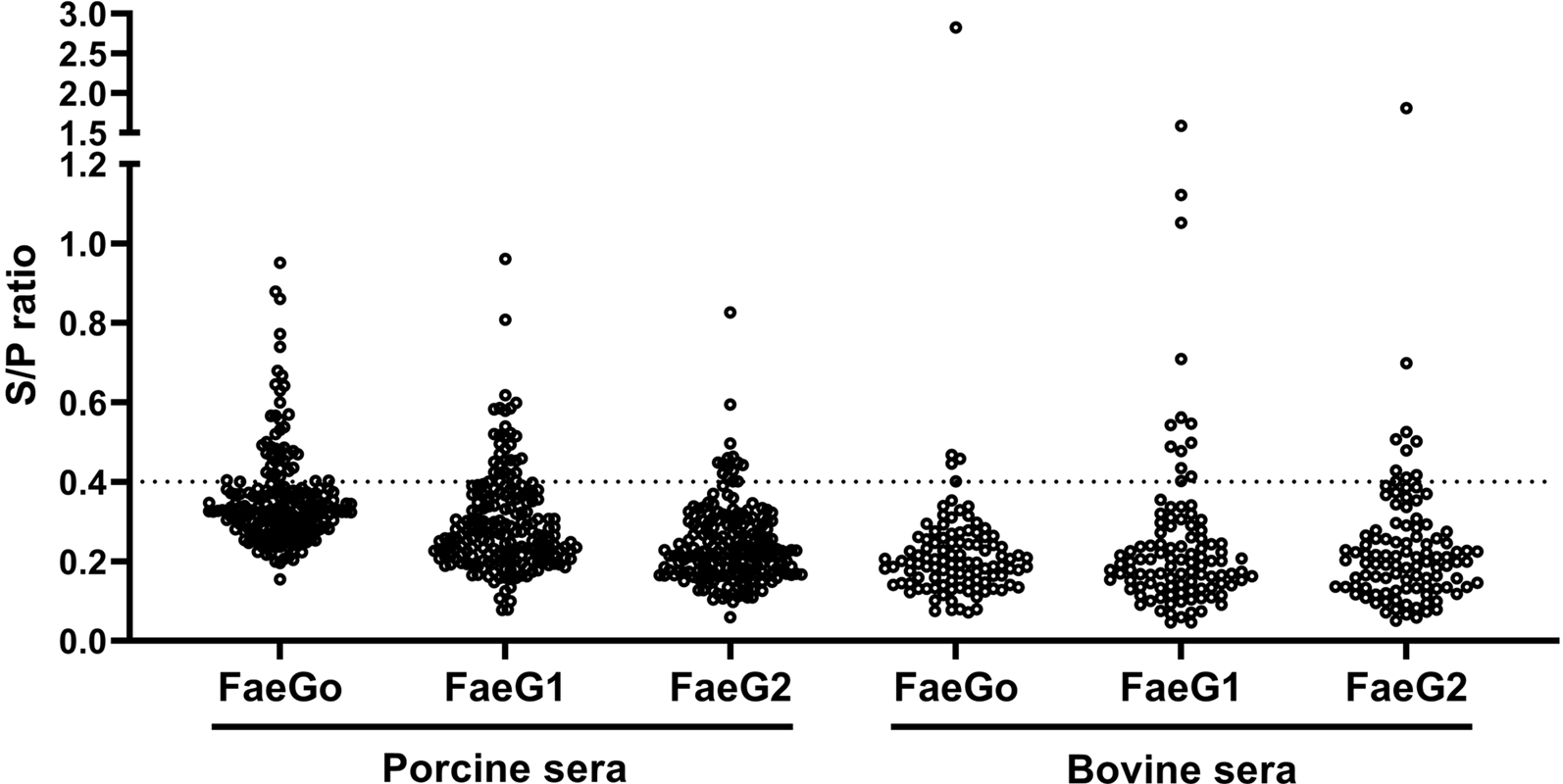
Fig. 1. Indirect ELISA plot of porcine and bovine sera. S/P ratio for individual porcine and bovine sera against FaeG1, FaeG2 and FaeGO.
Table 1. Prevalence of antibodies against FaeGo, FaeG1 and FaeG2 among porcine and bovine sera
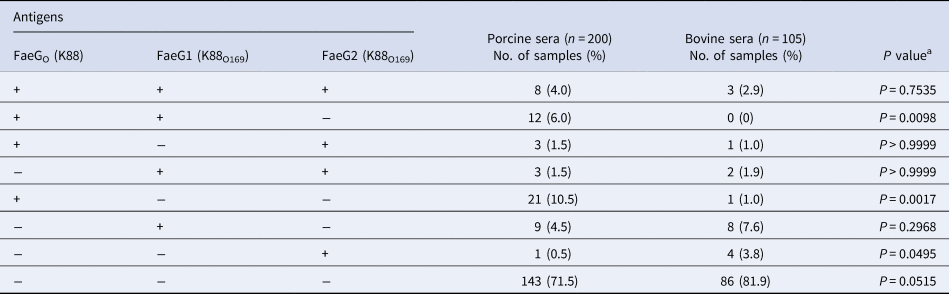
a Fisher's exact test was employed to assess the association between porcine and bovine seroprevalence.
There were no statistical differences in the prevalence of K88O169 antibodies reacting to FaeG1 and/or FaeG2 between porcine and bovine sera. Antibodies against FaeG1 were detected in 16.0% (32/200) of pigs and 12.4% (13/105) of cattle and anti-FaeG2 antibodies were found in 7.5% (15/200) of pigs and 9.5% (10/105) of cattle. There were no statistical differences in the prevalence of antibodies to each of FaeG1 and FaeG2 between porcine and bovine sera (FaeG1; P = 0.4979, FaeG2; P = 0.5199). Anti-FaeG1 single positive sera were higher for cattle (n = 8, 7.6%) than pigs (n = 9, 4.5%) (P = 0.2968) (Table 1). Anti-FaeG2 single positive sera were significantly higher for cattle (n = 4, 3.8%; P = 0.0495) than pigs (n = 1, 0.5%). In contrast, evaluation of the prevalence of antibodies against FaeG of K88ac (hereinafter, FaeGO, to reflect its originality), a typical adhesion factor of porcine ETEC, shows that the antibodies to FaeGO were markedly more prevalent among pigs than cattle (Table 1, Fig. 1).
Comparison of epitopes of FaeGO, FaeG1 and FaeG2 adhesins shows differences in amino acid sequences (Fig. 2) [Reference Lu, Moxley and Zhang15]. Individuals positive for antibodies to FaeGO of K88 did not necessarily cross-react to FaeG1 or FaeG2 of K88O169 (Fig. 3). The presence of antibodies to FaeG1 was not associated with the presence of antibodies to FaeG2, and vice versa (Fig. 3). A total of 27 samples were positive for FaeG1 or FaeG2 antigens but not FaeGO, and 22 anti-FaeGO positive samples were negative for FaeG1 and FaeG2 antigens (Table 1).

Fig. 2. Epitope comparison of FaeGO, FaeG1 and FaeG2. The amino acid sequences of FaeGO, FaeG1 and FaeG2 were aligned using ClustalW. Residues identical to FaeGO are shown as dots. Hyphens indicate positions where residues are missing. The residue regions enclosed in rectangles are the epitopes referenced in a previous study [Reference Lu, Moxley and Zhang15].
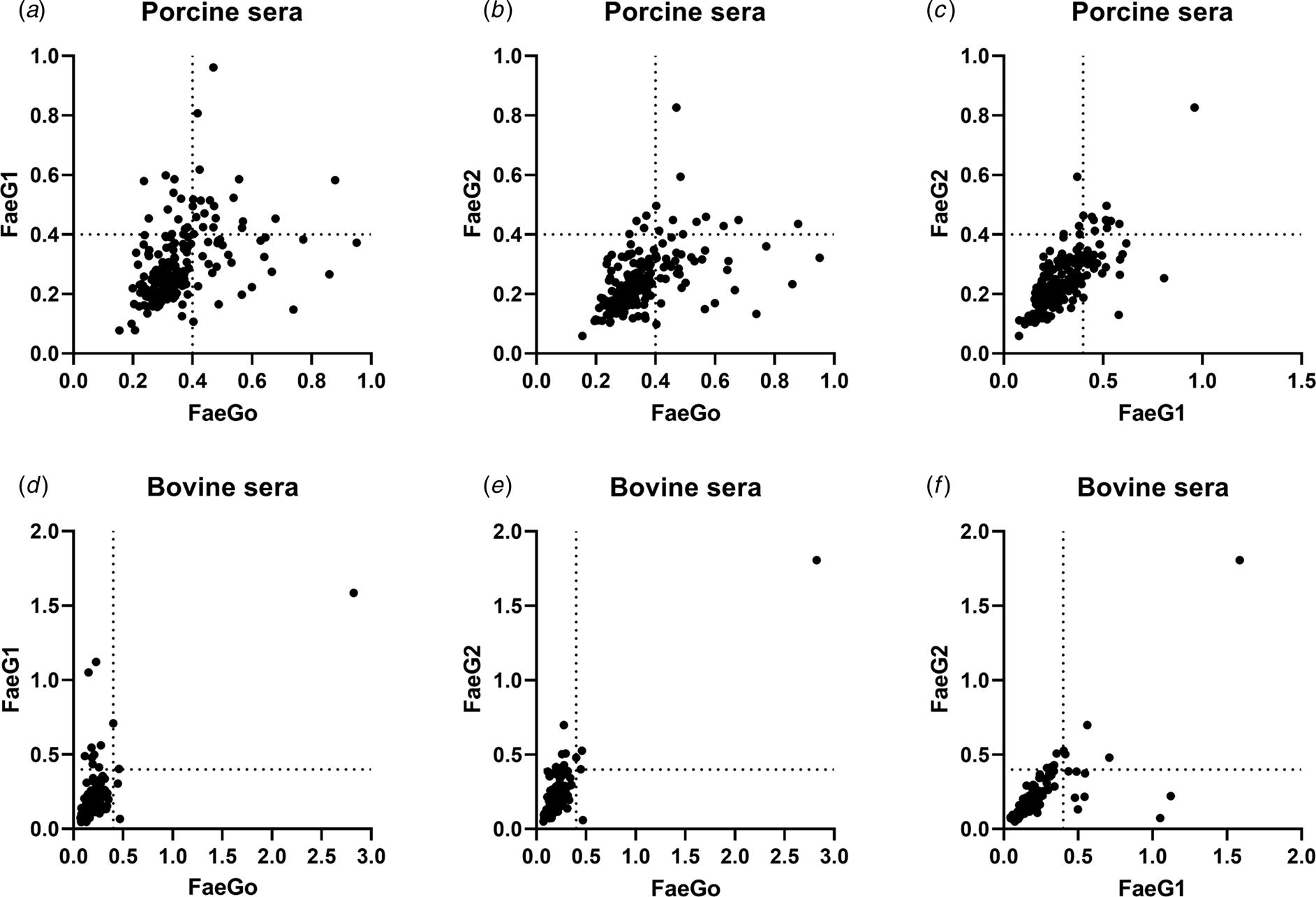
Fig. 3. Indirect ELISA plots of FaeG1 or FaeG2 against FaeGO. Antigens of FaeG1 or FaeG2 plotted against FaeGO for porcine (a and b) and bovine sera (d and e) show the absence of cross-reaction between each of two antigens. Antibody titres to FaeG1 and FaeG2 showed no relationship to each other in porcine and bovine sera (c and f).
Discussion
The O169 plasmid pEntYN10 has a total of three CFs (CS6, K88-like and CFA/III-like) [Reference Ban6]. CS6 is recognised as being one of the representative CFs for humans; however, the role of the other two CFs had not yet been elucidated. Since the O169 plasmid can easily be eliminated from bacteria in vitro [Reference Ban6], the presence of multiple CFs in the plasmid suggests the possibility that the bacteria can infect various species of hosts consecutively. We used seroepidemiological screening to evaluate whether ETEC possessing K88O169 could infect pigs because K88 is a classical CF of porcine ETEC.
Porcine and bovine sera reacted to one or both of FaeG1 and FaeG2 antigens of K88O169. These results suggest that K88O169-positive E. coli might infect pigs and cattle. Although the K88O169 operon codes two paralogous major adhesin-like subunits, FaeG1 and FaeG2, FaeG1 seems likely to be the adhesin since anti-FaeG1 was obviously more prevalent than anti-FaeG2. The FaeGO positivity rate was significantly higher among pigs than cattle (P < 0.0001), which seroepidemiologically reflects that colonisation of ETEC possessing K88 is more prevalent among pigs. This is consistent with previous reports that the original K88 is a typical adhesion factor of ETEC that causes diarrhoea in piglets [Reference Woodward16, Reference Luppi17]. K88ac was used as a positive control antigen, and we consider that these data support the validity of using ELISA in this study.
We considered the possibility that anti-FaeGO cross-reacts with FaeG1 and FaeG2. However, epitopes of FaeGO, FaeG1 and FaeG2 adhesins show differences in amino acid sequences. Individuals positive for antibodies to FaeGO, FaeG1, or FaeG2 alone were observed in both pigs and cattle. These results suggest that individuals infected with the bacteria that harbour FaeG1, FaeG2, or the homologous CF could be prevalent in these species; positive reactions to these novel antigens are unlikely due to the cross-reaction of antibodies to FaeGO.
CF is generally recognised as being the mechanism that determines host specificity. However, our findings showed that anti-K88O169 was prevalent among bovine sera and porcine sera, suggesting that K88O169 may provide the strain with a mechanism that allows it to infect multiple hosts. The presence of CS8-like CF plasmid, the third and novel one of the O169 strain, may also enhance survival strategies by expanding the host spectrum bacteria in which it is present. Interestingly, virulence plasmid pEntYN10 tended to be quickly eliminated in vitro because there are fewer genes associated with plasmid maintenance [Reference Ban6]. In contrast, this plasmid is maintained in ETEC O169 despite this disadvantage; the plasmid likely includes CFs that make it possible for the bacteria to infect multiple hosts, which, in turn, allows the bacterial hosts to retain the plasmid.
Seroepidemiology is an indirect detection method, and the detection of antibodies can not be used to prove that the animal was infected with or that it ever carried the infectious agent being studied. We have not yet isolated these types of bacteria from animals and livestock products. Further, we do not know if the K88O169-possessing bacteria are pathogenic or innocuous in animals; specific hosts where the K88O169 acts as the CF remain to be identified. Another investigation is ongoing to look for K88O169-possessing bacteria in the porcine and bovine faecal matter by PCR using primer sets designed for the genes of the K88O169 family.
In conclusion, antibodies to K88O169 antigens are prevalent among pigs and cattle. K88O169-possessing organisms may expand their niches through a unique and complex repertory of CFs. The present data imply that not only O169 but also other serogroups of E. coli possessing K88O169 could be transmitted from domestic animals to humans or from humans to domestic animals through the switchover of different adhesins across hosts. We should pay attention to ETEC as a possible pathogen of zoonosis.
Supplementary material
The supplementary material for this article can be found at https://doi.org/10.1017/S0950268821002697
Acknowledgements
The authors thank veterinarians Mitsuhiro Tsujimoto, Tomofumi Maehara, Norihide Kuriyama, Hideki Oshima and Yusuke Kataoka, at the Osaka City Meat Hygiene Inspection Center, for providing porcine and bovine sera.
Author contributions
YT and YN designed the study. YT, MI, KK and AO carried out the experiments. YT analysed data. EK-N was involved in planning and supervising the work. YT, TW and YN wrote the paper.
Financial support
This work was supported by a Grant-in-Aid for Scientific Research (B) (17H04078), Grant-in-Aid for Early-Career Scientists (19K16639) and Grant-in-Aid for Exploratory Research (19K22459) from the Japan Society for the Promotion of Science (JSPS).
Conflict of interest
The authors declare no competing interests.
Data availability statement
The datasets generated during the current study are available from the corresponding author on reasonable request.







