Since their earliest descriptions, major mental illnesses such as schizophrenia and bipolar disorder have been known to run in families. We now know from family, twin and adoption studies that they have a strong genetic component and that genes contribute substantially to their pathogenesis (Reference Sullivan, Kendler and NealeSullivan 2003; Reference Lichtenstein, Yip and BjörkLichtenstein 2009). Initial attempts to find the responsible genes, based on genetic linkage analysis and candidate gene association studies, met with limited success and lack of replication. However, the field has advanced considerably over the past few years, with the advent of genome-wide association technologies and large international collaborations. This has led to major advances in identifying genetic risk variants for major psychiatric conditions. Among the most exciting and important of these findings has been the discovery of copy number variants (CNVs) and their role in psychiatric disorders.
What are CNVs?
Copy number variants are submicroscopic structural variations affecting more than 1000 DNA base pairs (Reference Lee and SchererLee 2010). They represent an important source of genetic diversity, with small, benign CNVs being found in the genomes of healthy individuals (Reference Iafrate, Feuk and RiveraIafrate 2004; Reference Sebat, Lakshmi and TrogeSebat 2004). However, numerous pathogenic CNVs have now been identified (Reference Girirajan, Rosenfeld and CoeGirirajan 2012; Reference Vissers and StankiewiczVissers 2012), such as those causing Prader–Willi and Angelman syndromes (deletions at 15q11–q13) and Williams–Beuren syndrome (deletions at 7q11.23) (Reference Stankiewicz and LupskiStankiewicz 2010). Syndromes caused by CNVs have consequently come to be called genomic disorders (Reference LupskiLupski 1998). Several classes of CNV are known to exist, each causing a different type of chromosomal structural change. These include balanced CNVs, such as inversions or translocations, that respectively change the orientation or location of a DNA segment (Reference Weischenfeldt, Symmons and SpitzWeischenfeldt 2013). Unbalanced CNVs involve either deletion or duplication of segments of DNA, accordingly reducing or increasing the usual number of copies of the segment (Reference Weischenfeldt, Symmons and SpitzWeischenfeldt 2013). For the remainder of this article, we will use CNV to refer to deletions and duplications (the unbalanced type: Fig. 1).
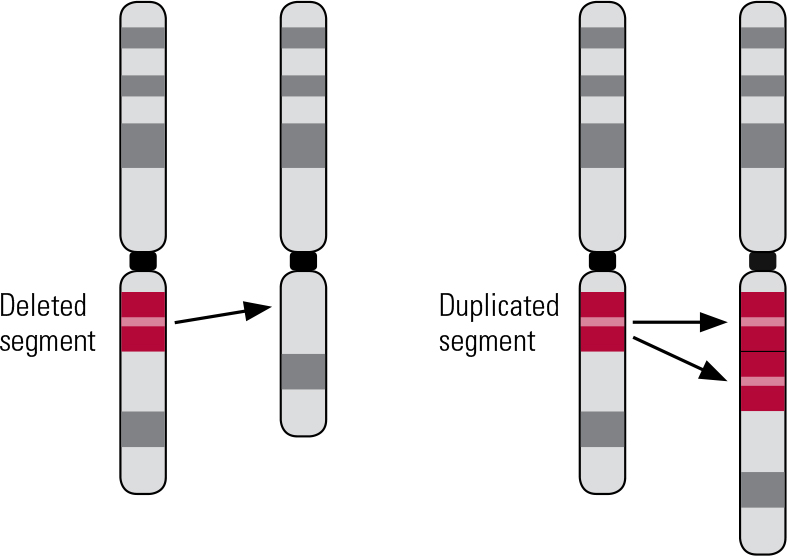
FIG 1 Schematic of a deletion and duplication.
Mechanisms of CNV formation
The majority of known pathogenic CNVs are recurrent, meaning that they occur in the same genomic locations (rather than randomly across the genome). The boundaries of recurrent CNVs tend to cluster in genomic regions that display high homology, typically regions of highly repetitive DNA sequences, such as low copy repeats (LCRs). These regions encourage CNVs to form by causing chromosomes to misalign during recombination (the crossing over of chromosomes that occurs during meiosis and mitosis). This mechanism is known as non-allelic homologous recombination (NAHR) (Reference Malhotra and SebatMalhotra 2012) (Fig. 2). NAHR can occur during meiosis, forming de novo (new) CNVs, or during mitosis, forming somatic CNVs (present in only a proportion of cells, or restricted to one tissue/organ of the body, where the mutation occurred post-conception) (Reference Simmons, Carvalho and LupskiSimmons 2012).
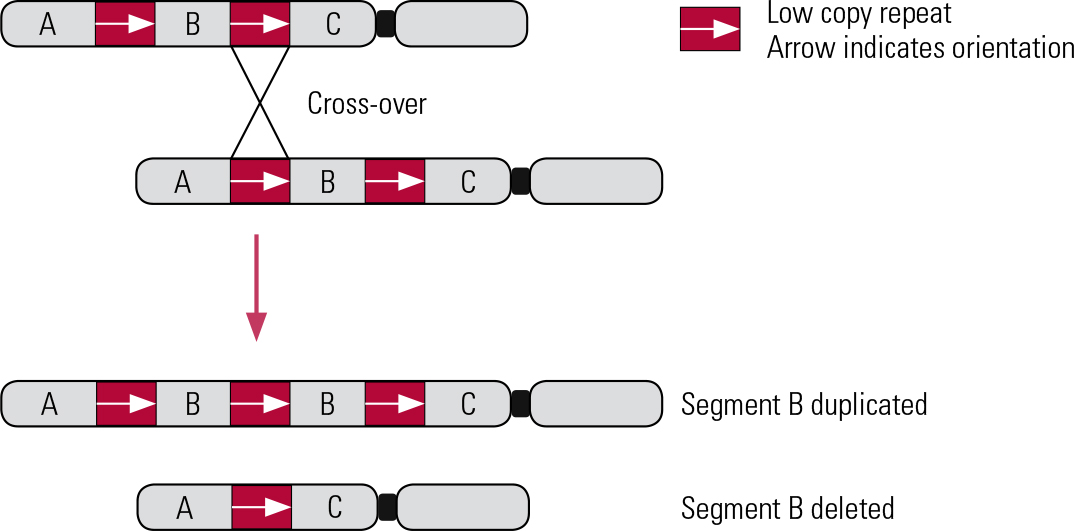
FIG 2 Non-allelic homologous recombination. Inter-chromosomal deletion and duplication. Red boxes and arrows indicate the location and orientation of low copy repeats (LCRs).
Human genomes also carry non-recurrent CNVs, which are more randomly distributed across the genome and have breakpoints at unique genomic locations (Reference Gu, Zhang and LupskiGu 2008). These CNVs are known to arise through errors in mechanisms involved in DNA break repair (e.g. non-homologous end joining) and DNA replication (e.g. fork stalling and template switching) (Reference Malhotra and SebatMalhotra 2012).
The estimated mutation rate of several CNV loci known to cause genomic disorders is high, affecting between 1:4000 and 1:25 000 newborns (Reference LupskiLupski 2007). Very similar mutation rates have also been estimated for the CNV loci associated with schizophrenia (Reference Rees, Moskvina and OwenRees 2011).
CNVs in schizophrenia
For many years, the only genetic factor known to increase risk for schizophrenia was a deletion of about 3 million base pairs on chromosome 22q11.2, which is a CNV flanked by several blocks of LCRs and deletes over 40 genes. In 1978 it was identified as causing a severe genetic disorder, variably called velocardiofacial syndrome (VCFS), DiGeorge syndrome or Shprintzen syndrome (Reference Shprintzen, Goldberg and LewinShprintzen 1978). It was known to occur as a new mutation in ~1:4000 births, and ~30% of carriers of that deletion developed a psychotic disorder (Reference Murphy, Jones and OwenMurphy 1999). Carriers of the deletion have a reduced IQ of ~70 and a number of physical anomalies, such as cleft lip/palate, heart defects and facial changes.
It took many years before further discoveries were made, enabled by the rapid development of new technologies that allowed the interrogation of the entire human genome on a single chip (microarray), initially with ~30 000 probes, and more recently with ~500 000 to 2000 000 probes (Fig. 3). Some of these arrays were designed for the genotyping of single nucleotide polymorphisms (SNPs), but it became obvious that they could also identify the number of copies of genetic material (i.e. deletions or duplications) at these sites, by using the signal intensities of the probes.
FIG 3 Examples of microarrays: (a) Affymetrix® and (b) Illumina® arrays.
The year 2008 saw the first two landmark papers (International Schizophrenia Consortium 2008; Reference Stefansson, Rujescu and CichonStefansson 2008) that confirmed the role of VCFS mutations and identified three new deletions as risk factors for schizophrenia: at 1q21.1, 15q11.2 and 15q13.3. These were also large deletions of between 300 000 and 1 300 000 base pairs of genetic material, deleting between 4 and 11 genes. The new technologies allowed very rapid genotyping of thousands of samples, thus enabling the identification of further loci. It became evident that these pathogenic CNVs were all very rare, found in 1:200 to 1:1500 patients with schizophrenia, and much rarer in controls. This indicated the need to analyse very large samples of patients and controls in order to obtain the necessary statistical power to discover an association. As of today, over 12 000 patients and up to 80 000 controls have been genotyped using appropriate microarrays, enabling the confirmation and independent replication of 11 loci at a genome-wide level of statistical significance (reviewed in Reference Rees, Walters and GeorgievaRees 2014). These loci are listed in Table 1. We should point out that more loci have been strongly implicated, but at present they do not meet our strict criteria for the multiple testing correction of 20 000 individual genes (P <1.25 × 10−6) or of 120 genomic regions that are potentially prone to recurrent CNVs (P < 4.1 × 10−4). Some of them could also become established loci when larger samples are analysed. They include, for example, deletions at distal 16p11.2, at 16p12.1, 17p12 and 17q12, and CNVs at the genes TOP3B, PAK7, VIPR2 and SLC1A1.
TABLE 1 List of 11 copy number variants (CNVs) implicated in schizophrenia at genome-wide levels of statistical significance

With the exception of deletions at the NRXN1 gene, all CNVs confirmed to increase risk for schizophrenia are very large (300 000–3 000 000 base pairs) and span multiple genes (Table 1). Not surprisingly, such large and multigenic events have large effects on health, with odds ratios (ORs) ranging from 2 to >50 for developing schizophrenia (Table 1). They also lead to multiple phenotypes, as discussed later. The only single gene confirmed to increase risk when affected by CNVs is NRXN1. NRXN1 encodes for a presynaptic cell-adhesion protein, which binds with post-synaptic proteins called neuroligins and plays a vital role in the formation, maintenance and release of neurotransmitters in synapses in the brain.
The same CNVs increase risk for other early-onset developmental disorders
In parallel with these discoveries, other teams were working on elucidating the role of CNVs in children and young adults who had been referred for genetic testing because of early-onset developmental disorders such as intellectual disability, congenital malformations and autism spectrum disorders (ASD). Until a few years ago, genetic testing typically involved karyotyping (inspection of chromosomes under a microscope). This type of investigation could identify only very large chromosomal aberrations. With the use of the new arrays, there was the option to search for smaller CNVs, of the type found in schizophrenia. Two very large studies reported on CNVs in individuals referred for genetic testing for such disorders. Reference Girirajan, Rosenfeld and CoeGirirajan et al (2012) reported on 32 587 such individuals who had been referred to one genetic laboratory, and Reference Kaminsky, Kaul and PaschallKaminsky et al (2011) reported results gathered by a consortium of cytogenetic laboratories on another 15 749 individuals. The results were rather unexpected for the psychiatric community: the same CNVs shown to increase risk for schizophrenia were found also to increase risk for these early developmental disorders (Fig. 4 and Table 2). In most cases the frequencies of these CNVs were greater in people with these disorders than in those with schizophrenia (for reviews see: Reference Malhotra and SebatMalhotra 2012; Reference Kirov, Rees and WaltersKirov 2014). Figure 4 presents the reported CNV frequencies in control populations and individuals with schizophrenia or one of the early-onset developmental disorders based on data reported in Reference Kirov, Rees and WaltersKirov et al (2014).
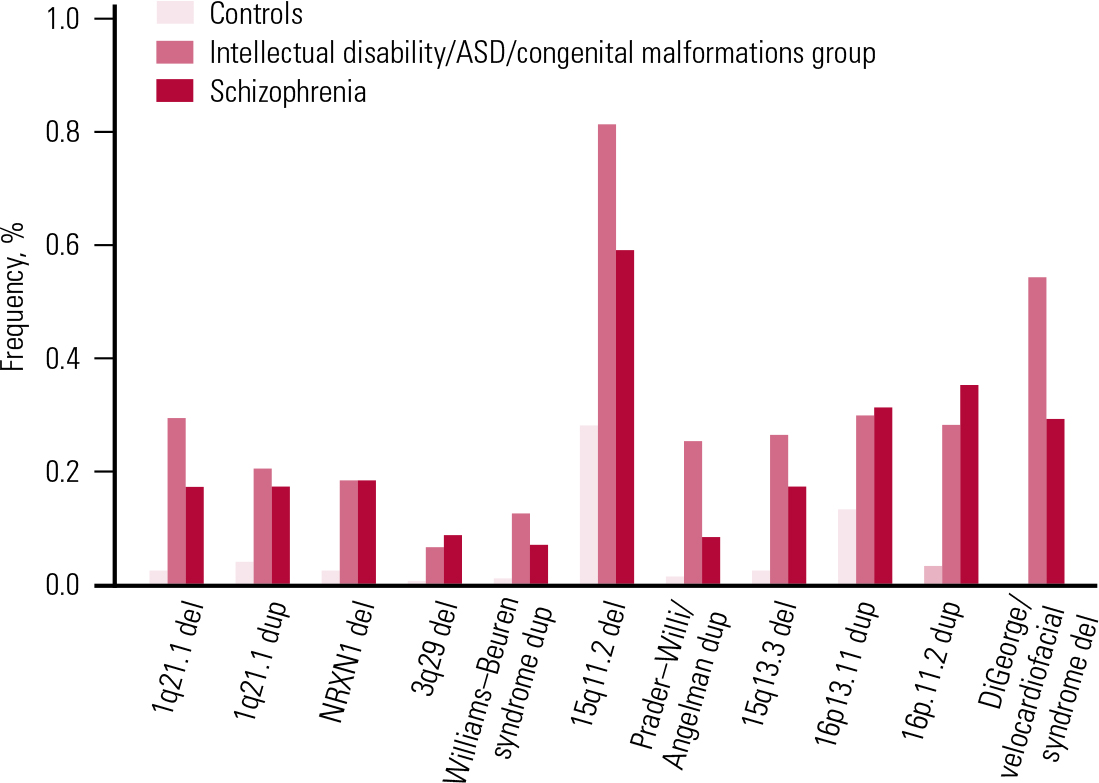
FIG 4 Frequencies of the most strongly implicated copy number variants in schizophrenia and early-onset developmental disorders. ASD, autism spectrum disorder; del, deletion; dup, duplication. Reprinted from Reference Kirov, Rees and WaltersKirov et al (2014), with permission from Elsevier.
TABLE 2 Frequencies and penetrance of CNVs
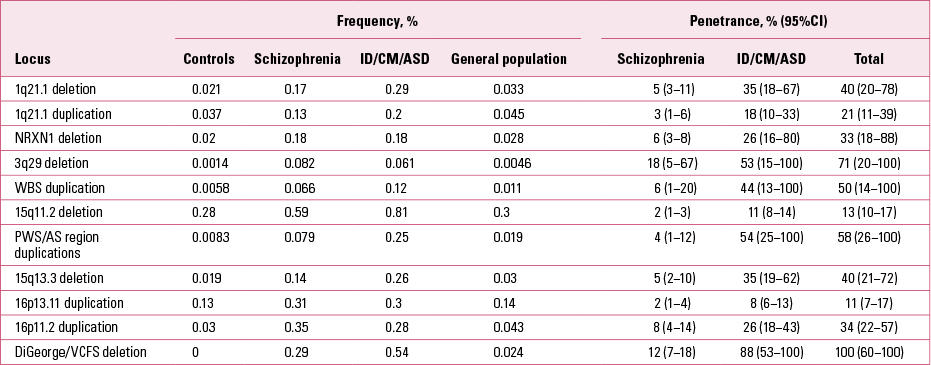
There are other CNV loci that specifically increase risk of intellectual disability, congenital mal formations or ASD, but are not found in schizophrenia (Reference Kirov, Rees and WaltersKirov 2014). They typically cause more severe and earlier-onset syndromal disorders, such as Williams–Beuren syndrome (deletions), Prader–Willi and Angelman syndromes (deletions) and Sotos syndrome (deletions). Cumulatively, pathogenic CNVs are found in ~7% of individuals with intellectual disability, congenital malformations or ASD and in 2.5% of people with schizophrenia (Reference Girirajan, Rosenfeld and CoeGirirajan 2012; Reference Rees, Walters and GeorgievaRees 2014). The rates of these CNVs in control populations are much lower (Table 1). The cumulative chance of observing a carrier of one of the 11 CNVs from Table 1 in an unscreened control is only 0.5% and is likely to be lower if controls are screened for health and cognitive performance. In fact, a recent study found cognitive deficits in healthy carriers of these CNVs (Reference Stefansson, Meyer-Lindenberg and SteinbergStefansson 2014). Independent of the early-onset developmental disorders, the CNVs implicated in schizophrenia have also been shown to increase risk of other specific phenotypes, such as epilepsy (15q11.2 and 15q13.3), microcephaly (1q21.1, 3q29 and 16p11.2) and congenital heart defects (1q21.1 and 22q11.2) (Table 1). Individuals with ASD have also been recruited and studied separately (not as part of groups referred for genetic testing). Several of the CNVs implicated in schizophrenia were shown to be among the most frequent genetic factors that increase susceptibility for ASD: duplications at the Prader–Willi/Angelman syndrome region, 16p11.2 duplications and 22q11.2 deletions (Reference Sanders, Ercan-Sencicek and HusSanders 2011). These novel and largely unexpected findings indicated genetic and biological overlap between the aetiology of schizophrenia and autism.
Penetrance of CNVs for schizophrenia and other disorders
It is clinically important to know the risk for schizophrenia in carriers of these CNVs (i.e. the penetrance of the disorder – the percentage of carriers that will develop the disorder). Reference Vassos, Collier and HoldenVassos et al (2010) were the first to estimate the penetrance of a subset of the CNVs for schizophrenia listed in Table 1. They found modest rates: 2–7.4% (and a much higher rate of 55% for the VCFS deletion on 22q11.2). These modest levels of penetrance brought into question the potential utility of CNVs as clinical tools in genetic counselling. However, the above-mentioned studies of Reference Kaminsky, Kaul and PaschallKaminsky et al (2011) and Reference Girirajan, Rosenfeld and CoeGirirajan et al (2012) changed the picture. The penetrance of some of the CNVs for the early-onset developmental disorders (intellectual disability, congenital malformations, ASD) was estimated to be much stronger, at between 10.4% for 15q11.2 deletions and 46.8% for the 16p11.2 duplications (Reference Rosenfeld, Coe and EichlerRosenfeld 2013). The reasons for the higher estimates are twofold. First, the frequencies of most of these CNVs among such patients are even higher than in schizophrenia (Fig. 4). Second, the population prevalence of the combined group of intellectual disability, congenital malformations and ASD is several times higher than that of schizophrenia. This means that many more people will develop one of these early-onset disorders, i.e. the penetrance is much higher for an early neurodevelopmental disorder than for schizophrenia. It is also becoming clear that in any genetic counselling, it would be important to consider not only the risk for schizophrenia, but also the risk for any other severe neurodevelopmental disorder.
The combined penetrance of CNVs for neurodevelopmental disorders
We therefore decided to estimate the overall penetrance for such disorders among CNV carriers (Reference Kirov, Rees and WaltersKirov 2014). We approximated the lifetime risk for schizophrenia at 1%, and for the combined group of intellectual disability, congenital malformations and ASD at 4%, and used the frequencies of CNVs from Table 1 for schizophrenia and controls, and the rates reported by Reference Girirajan, Rosenfeld and CoeGirirajan et al (2012) for the combined group of early-onset developmental disorders. As expected, the resulting estimates for penetrance were much higher for the group of early-onset developmental disorders than for schizophrenia (Table 2 and Fig. 5). The overall penetrance of these CNVs ranges from 11% for duplications at 16p13.11 to 100% for the VCFS region (where no carriers in the control populations have been observed, i.e. all carriers in the tested populations ended up having a disease). These are very substantial increases in risk.
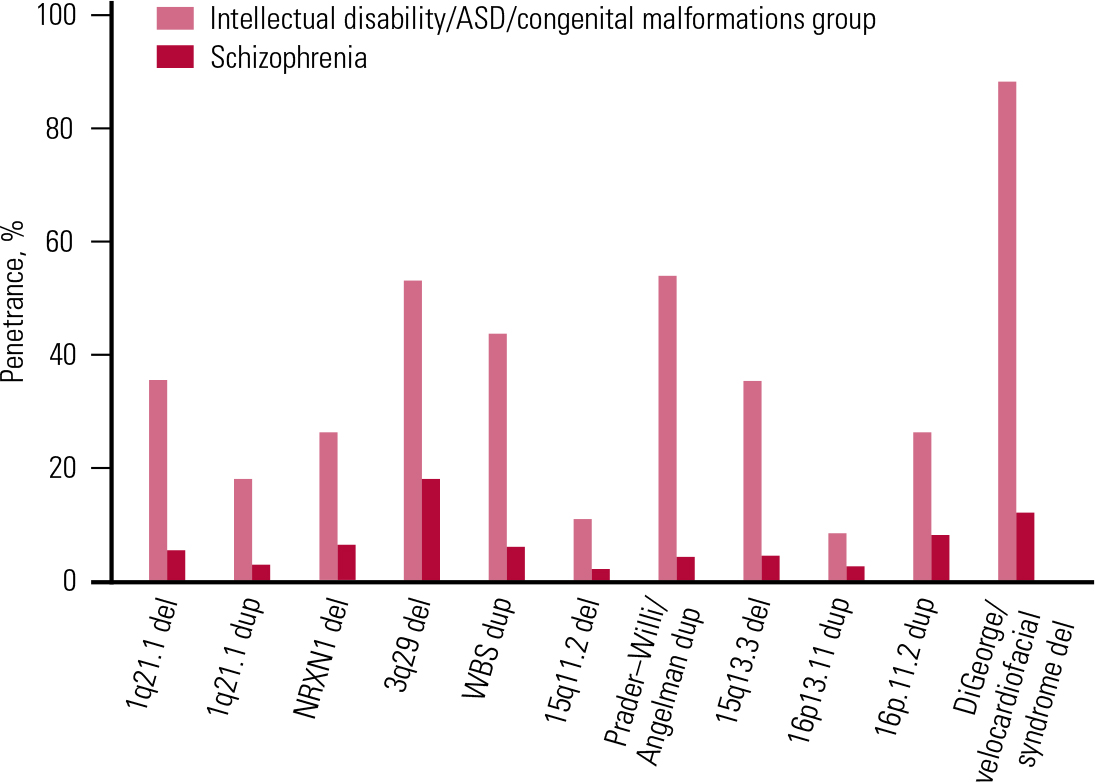
FIG 5 The penetrance of a selected set of copy number variants for schizophrenia and the group of intellectual disability, autism spectrum disorder (ASD) and congenital malformations. Reprinted from Reference Kirov, Rees and WaltersKirov et al (2014), with permission from Elsevier.
Selection against CNVs
People with schizophrenia tend to have fewer children, mainly owing to the impact that the condition has on forming and maintaining relationships. These trends have been shown in several large studies, most clearly those of large population-based samples from Denmark, Sweden and Finland (Reference Haukka, Suvisaari and LönnqvistHaukka 2003; Reference Laursen and Munk-OlsenLaursen 2010; Reference Power, Kyaga and UherPower 2013). For example, Reference Haukka, Suvisaari and LönnqvistHaukka et al (2003) demonstrated that, compared with the general population, women with schizophrenia have about half the number of children, and men with the disorder about a quarter the number. Individuals with intellectual disability, congenital malformations or ASD also have fewer offspring, although the exact figures will differ strongly according to the type of impairment. If carriers of a genetic variant are more likely to develop one of these disorders, such carriers are therefore also more likely to have fewer children. Indeed, this was recently demonstrated in the Icelandic population for carriers of some of these CNVs (Reference Stefansson, Meyer-Lindenberg and SteinbergStefansson 2014). As a consequence, there is a tendency for these genetic variants to become rarer in successive generations and eventually to be eliminated from the general population, i.e. they are under selection pressure.
CNVs have high mutation rates
The above observations raise the question of why the CNVs in Table 1 are still present in the general population. The obvious answer is that they arise at fairly high frequencies as de novo mutations in newborns. Indeed, we and others have shown that about 5% of people with schizophrenia have a de novo CNV (Reference Malhotra, McCarthy and MichaelsonMalhotra 2011; Reference Kirov, Pocklington and HolmansKirov 2012) and even higher rates of de novo CNVs have been reported in ASD and intellectual disability (Reference Sanders, Ercan-Sencicek and HusSanders 2011; Reference Rosenfeld, Coe and EichlerRosenfeld 2013). The mutation rates for most individual loci in Table 1 are indeed very high, ranging from 1:4000 to 1:26 000 newborns (Reference Rees, Moskvina and OwenRees 2011). This is because most of them are flanked by highly repetitive sequences of DNA that cause mis-pairing of DNA strands during mitosis and meiosis (Reference Malhotra and SebatMalhotra 2012).
Selection coefficients of CNVs
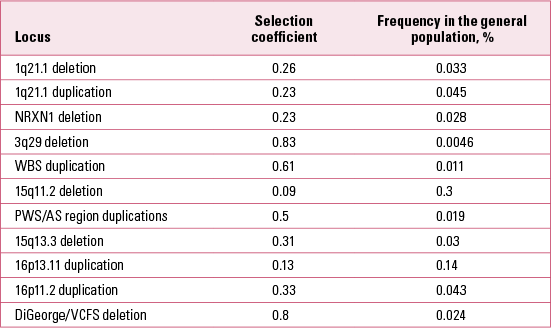
It is also possible to estimate the selection coefficients acting against carriers of the CNVs in Table 1 (Reference Rees, Moskvina and OwenRees 2011; Reference Kirov, Rees and WaltersKirov 2014). We estimated these after making the simplifying assumption that the frequency of each CNV in the general population remains about the same from generation to generation. Therefore the number of new mutations at a given locus in the general population is approximately equal to the number of CNVs eliminated through natural selection in one generation. We have data on the proportion of de novo CNVs out of the total number of CNVs at each locus in the general population. The selection coefficient (the proportion of CNVs eliminated in one generation due to natural selection) is thus estimated as the ratio of de novo CNVs to the total number of CNVs (de novo + those transmitted from parents to offspring). The results are presented in Table 3. These results indicate that all CNVs are under strong selection pressure, ranging from 0.09 to 0.83. Owing to the high selection pressure against these CNVs, each new mutation is eliminated from the general population within a very small number of generations, typically only 1–5 on average (Reference Rees, Moskvina and OwenRees 2011). The rate of these CNVs in the general population is therefore maintained by the balance between new mutations and the selection pressure operating against them.
TABLE 3 Selection coefficients of copy number variants
Implications for genetic counselling and testing
The accumulated knowledge about these genetic factors that have such large effect raises the question of their usefulness for genetic counselling and testing. Approximately 2.4% of people with schizophrenia carry one of these 11 pathogenic CNVs (Reference Rees, Walters and GeorgievaRees 2014). Apart from schizophrenia, these CNVs can cause intellectual disability and ASD and a range of other medical conditions, including epilepsy, congenital heart disease and microcephaly. Such comorbid conditions, and/ or the presence of pronounced cognitive deficit in a patient, should raise the clinician’s suspicion that the individual could be a carrier of one of the pathogenic CNVs. For these reasons, CNV testing is becoming routine in some neurodevelopmental clinics, the issue is beginning to receive attention in the literature (Reference Gershon and Alliey-RodriguezGershon 2013) and there are advocates of rolling out such testing to relevant psychiatric populations. The possibility of expanding testing to psychiatric clinics raises important practical and ethical issues and has wide implications for the current and future practice of psychiatry. There is limited research into the possible benefits and risks of such genetic testing and we would strongly advocate for such research. The increasing popularity of direct-to-consumer genetic testing makes such research efforts all the more important. Genetic questions are likely to be raised with increasing frequency in psychiatric clinics, given advances in public understanding of genetics and replicated findings confirming the substantial role genetics plays in neuropsychiatric conditions. It will be important to have an informed psychiatric profession that is genetically literate, if psychiatrists are to serve patients in this respect and contribute to the debate about CNV and wider genetic testing.
MCQs
Select the single best option for each question stem
-
1 In patients with schizophrenia you would expect to find a relevant CNV in:
-
a 1:1000
-
b 1%
-
c 2–3%
-
d 5–10%
-
e 1:4000.
-
-
2 Carriers of the 1q21.1 deletion are more likely to have:
-
a increased weight
-
b average IQ
-
c epilepsy
-
d heart defects
-
e diabetes.
-
-
3 The CNVs that increase risk of developing schizophrenia:
-
a are nearly always inherited from affected parents
-
b occur as de novo mutations in about 1:100 people from the general population
-
c are associated with fewer offspring in their carriers
-
d have a similar penetrance for the development of autism or intellectual disability
-
e double the risk of developing schizophrenia.
-
-
4 As regards CNVs associated with neuro-developmental disorders:
-
a they are common genetic variants of small effect
-
b they are randomly distributed across the genome
-
c all carriers of such CNVs will have comorbid physical problems
-
d all carriers of large, rare CNVs of this sort will have psychiatric problems
-
e they are rare and of large effect size.
-
-
5 As regards the nature of CNVs (relevant to genetic counselling):
-
a carriers of CNVs associated with neuro-developmental disorders usually cannot have children
-
b de novo CNVs are inherited from a parent
-
c a CNV carrier has a 25% chance of passing the CNV onto their children
-
d a minority of VCFS (22q11.2) deletions are inherited
-
e CNVs associated with neurodevelopmental disorders are typically inherited from both parents who are carriers.
-
MCQ answers
| 1 | C | 2 | d | 3 | c | 4 | e | 5 | d |











eLetters
No eLetters have been published for this article.