INTRODUCTION
Head and neck squamous cell carcinoma (HNSCC) is classified as the seventh most common cancer worldwide with around 680 000 new diagnoses each year [Reference Ferlay1]. While tobacco and alcohol use are the principal risk factors that have been identified for head and neck cancer, human papilloma virus (HPV) has been aetiologically associated with 20–60% of HNSCC cases, particularly cases involving the oropharynx [Reference Kreimer2]. The incidence of HPV-correlated oropharyngeal cancers has increased over the past few decades, particularly in males. In addition, several studies have reported that individuals infected with human immunodeficiency virus (HIV) have an increased risk of developing HNSCC, potentially due to the effects of HIV-related immunosuppression [Reference Gillison3, Reference Denny4].
HPV-16 has been detected in ~30% of HNSCC cases, followed by HPV-18 (8·0%) [Reference Ferlay1]. While the most common genotypes in HPV-related HNSCC cases have been reported, information regarding the patterns of oral HPV genotypes in individuals without cancer or with pre-cancer lesions are lacking. Some studies suggest that the prevalence of HPV oropharyngeal infections (0·9–7·5%) [Reference Chung, Bagheri and D'Souza5] is substantially lower than that of HPV genital infections in healthy individuals, yet the prevalence of HPV oropharyngeal infections is still much higher in HIV-infected subjects [Reference Beachler6], particularly for men who have sex with men (MSM) [Reference Mooij7]. Moreover, limited data are available regarding the distribution of oropharyngeal HPV types in MSM. Greater knowledge of the HPV oropharyngeal infections that affect the MSM population could have important public health implications, especially for the identification of possible preventive options for HPV-related head and neck cancer. HPV screening and HPV vaccination are two of the preventive options that are currently being considered.
The aim of this study was to investigate the epidemiological profile of HPV oropharyngeal infections in HIV-infected MSM. Therefore, the pattern of oropharyngeal HPV genotypes circulating in an Italian cohort of HIV-positive MSM is reported, along with the prevalence, incidence, and persistence of oropharyngeal HPV infections in this population.
METHODS
Study design and sample collection
Between January 2011 and October 2012, 135 HIV-positive MSM that consecutively visited the Sexually Transmitted Diseases (STD) Outpatient Clinic of the L. Sacco University Hospital (Milan, Italy) for routine HIV infection monitoring were enrolled in this study. Each participant was asked to return for a follow-up control visit at least 12 months later and 70 (51·9%) subjects have attended the visit. At each visit, an oropharyngeal swab sample was collected from each participant. Briefly, for each sample, vigorous scraping of the walls of the oropharyngeal cavity with a swab that was rotated twice clockwise and twice anticlockwise was performed. The swabs were subsequently preserved in sterile tubes containing 3 ml phosphate-buffered saline (PBS), and stored at −20 °C until undergoing testing at the Virology Laboratories of the Department of Biomedical Sciences for Health, University of Milan (Milan, Italy).
HPV DNA detection
To obtain the cellular component of each swab sample, 1·5 ml of each sample received 3 h centrifugation at 16 000 g. The entire pellet obtained was dissolved in 500 µl PBS and then stored at −20 °C until nucleic acid isolation was performed.
DNA was extracted from 500 µl oropharyngeal sample with a NucliSENS® easyMAG® kit (bioMérieux bv, France). The concentration and purity of the extracted DNA was evaluated with a spectrophotometer (NanoDrop ND-2000/2000C, Thermo Fisher Scientific Inc., USA).
HPV DNA was detected starting from 10 µl extracted DNA and using a specific nested polymerase chain reaction (PCR) assay that amplified a 150 base pair (bp) segment of the L1 open reading frame using two primer pairs: ELSI_F/ELSI_R and GP5+/GP6+ [Reference Tanzi8, Reference Qu9]. Positive (HPV-16-positive cells, Caski) and negative (water) control samples were included in each PCR assay. DNA integrity was assessed with amplification of a 268-bp fragment from the ubiquitous β-globin gene [Reference Puranen10]. Amplified PCR products were subjected to electrophoresis in a 2% agarose gel containing ethidium bromide (0·5 µg/ml) and were compared with DNA Molecular Weight Marker 100 (Sigma-Aldrich, USA). To validate the reproducibility of this HPV detection method the test was repeated twice for each sample.
HPV DNA sequencing and genotyping
All amplified HPV DNA fragments were sequenced using the standard Sanger method, in order to identify the broadest spectrum of genotypes able to infect the oral mucosa. This method allows characterization of the predominant sequence in the sample, thereby preventing potential co-infections to be detected.
Briefly, PCR products ~150 bp in length obtained from the nested PCR assays were purified using the NucleoSpin® Extract II kit (Macherey-Nagel GmbH, Germany), these were then sequenced with an ABI PRISM 3100 genetic analyser (Applied Biosystem, USA). L1 gene nucleotide sequences were aligned with the reference sequences in GenBank and Papillomavirus Episteme (https://pave.niaid.nih.gov) using the public bioinformatics program BLAST [11, 12] for viral genotype identification.
The International Agency for Research on Cancer (IARC) has assigned 25 HPV types to a high-risk clade (HR clade), and these have been subdivided into three groups [13]. Group 1 includes HPV types 16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, and 59, and these are considered ‘carcinogenic to humans’. The genotypes of the other two groups, 2A and 2B, are designated as ‘probably carcinogenic to humans’ and ‘possibly carcinogenic to humans’, respectively [13].
Statistical analysis
Prevalence of HPV infection was calculated as the proportion of HPV-positive subjects of the total number of enrolled patients. An incident infection at follow-up was defined as the detection of HPV DNA in a subject HPV negative at baseline. A cleared infection at follow-up was defined as undetectable HPV DNA in a previously HPV-positive subject. A persistent HPV infection was defined as detection of the same HPV genotype at both baseline and follow-up.
Data are presented as the median [interquartile range (IQR)] and percentage [with 95% confidence interval (CI)] as appropriate. Comparisons between groups were made using the χ 2 test or the mid-P exact test. A P value <0·05 was considered statistically significant (two-tailed test). All statistical analyses were performed using OpenEpi software, v. 3.03a [Reference Dean, Sullivan and Soe14].
Ethical statement
The study protocol used by the research units involved was approved by the Ethics Committee of L. Sacco University Hospital, Milan, Italy. Written informed consent was obtained from all participants. At enrolment, demographic data and information regarding the HIV status of each patient were collected.
RESULTS
Spectrum of HPV genotypes infecting the oropharyngeal mucosa
A total of 205 (135 and 70, at baseline and follow-up, respectively) oropharyngeal swabs were collected from HIV-infected MSM and analysed for HPV DNA. Fifty-one out of 205 samples were HPV DNA positive. The reproducibility of the HPV DNA detection method was 100%, since the same positive or negative result was obtained by repeating each sample twice.
Forty-three out of 51 samples were suitable for DNA sequencing. Twenty-two distinct HPV genotypes were identified (Fig. 1). HPV-16 and HPV-33 were most often identified in HR group 1 genotypes, and HPV-70 in group 2B. HPV-6 and HPV-32 were the most frequent among the low-risk (LR) genotypes. Of particular interest was the detection of two recently identified alpha-HPV genotypes, i.e. HPV-102 and HPV-114.
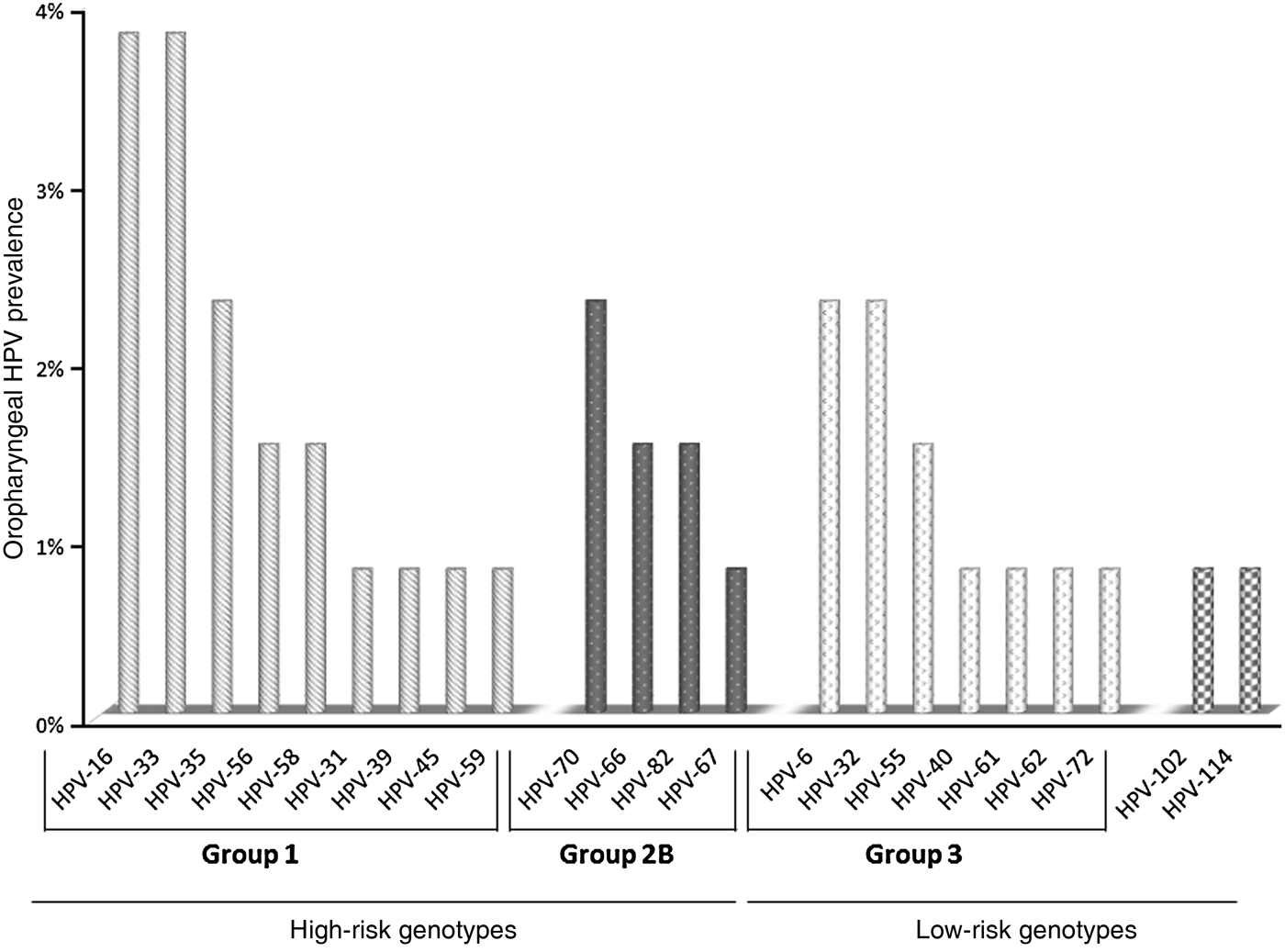
Fig. 1. HPV genotypes identified from oropharyngeal swab samples collected from the present cohort.
Epidemiological profile of oropharyngeal HPV infections in the study population
Demographic and clinical characteristics of the study population
The HIV-positive MSM included in this study (N = 135) ranged in age from 23 to 70 years (median 43, IQR 36–49); 115 (85·2%) were of Italian origin, whereas the remaining 20 (14·8%) were foreign subjects that were legally residing in Italy. Most patients (84·5%) were receiving combination antiretroviral therapy and had an undetectable HIV viral load (72·1%) and a CD4 cell count >500 cells/μl (55·9%, median 518 cells/μl).
Prevalence of oropharyngeal HPV infection
At baseline, 33/135 subjects (24·4%, 95% CI 17·8–32·2) were infected with HPV. The median age of the HPV-positive subjects was 42 years (IQR 36–51) and the 35–40 years age group was the most frequently infected, particularly by HR clade types (Fig. 2). All HPV-positive patients had a CD4 cell count >200 cells/μl (median 506 cells/μl, IQR 433-574 cells/μl).
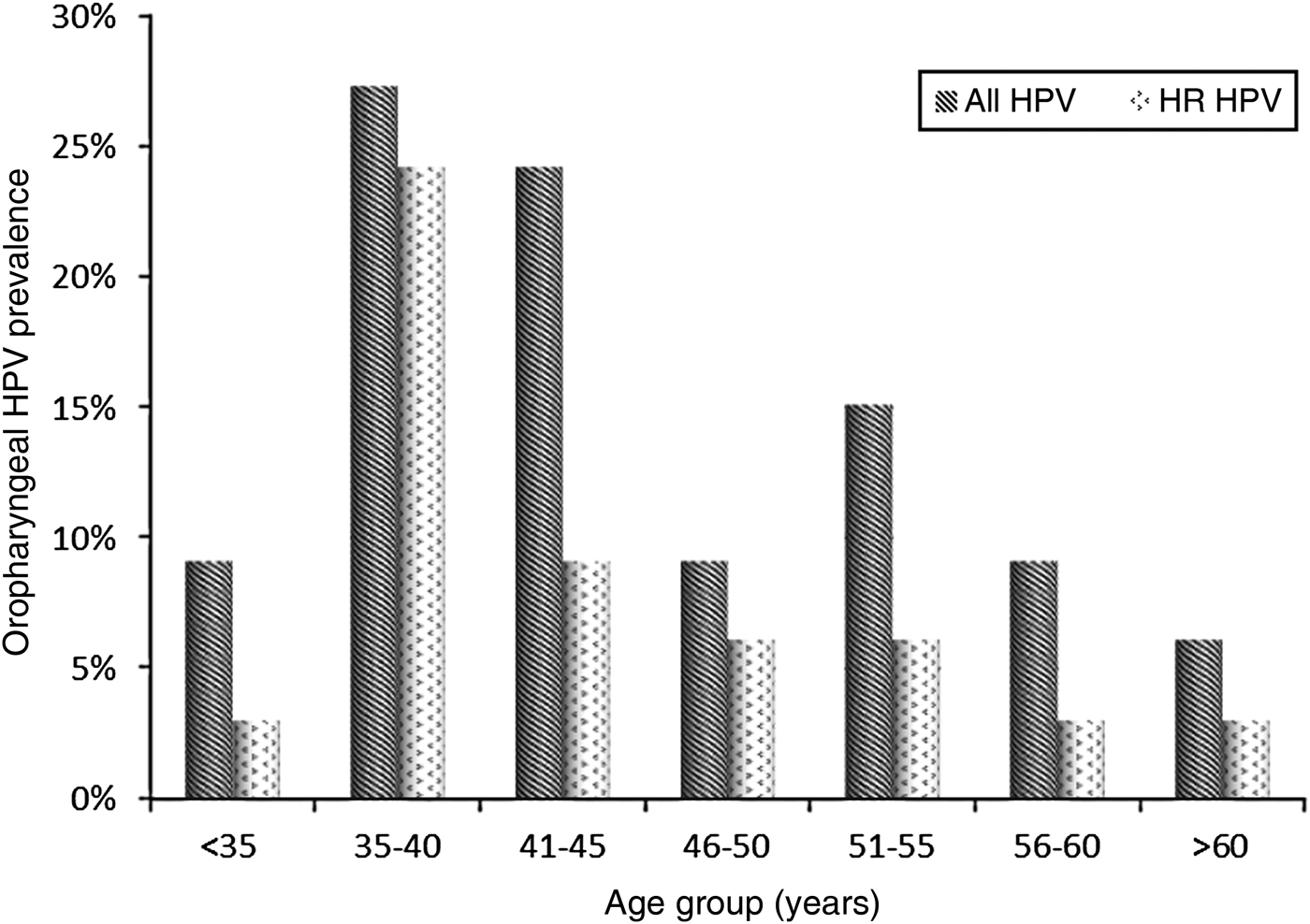
Fig. 2. Distribution of HPV-positive and high-risk (HR) clade HPV-positive MSM by age group.
The prevalence of infections sustained by HR HPV types was higher than that by LR HPV types (13·7% vs. 6·9%, respectively, P < 0·05) (Table 1). Of the HR HPV infections, HPV-16 and HPV-33 were the most common, each identified in 3·8% of the study cohort; infections with HPV-6 and HPV-32 were the most prevalent (2·3% each) in LR HPV infections. Two subjects were infected with two uncommon and recently identified alpha-HPV genotypes, respectively HPV-102 and HPV-114. These two genotypes had not been reported previously in oral mucosa.
Table 1. Prevalence of high-risk and low-risk HPV infections at baseline and follow-up
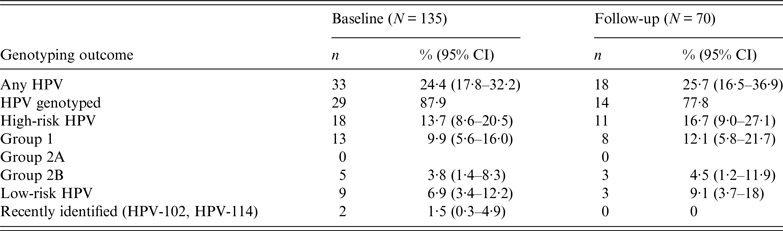
CI, Confidence interval.
Incidence and clearance of oropharyngeal HPV infection
Seventy out of 135 subjects (51·9%) returned at the follow-up visit at least 12 months later (median 18·5 months, IQR 12–29). Of these 70 subjects, 12 (17·1%) were positive for HPV DNA at baseline and 58 (82·9%) were negative. Of the latter, 14 subjects developed an HPV infection during the follow-up period, with an overall cumulative incidence of 24·1% (95% CI 14·5–36·4). The incidence of HR-HPV infections was 15·5%, mainly sustained by HPV-33, HPV-35, and HPV-70.
Of the 12 subjects HPV DNA positive at baseline, seven cleared the infection, with an overall infection clearance of 58·3% (95% CI 46·8–91·1). Only one subject (8·3%) had a HPV infection that persisted during the follow-up period, and was sustained by HPV-33. Finally, four subjects (33·3%), exhibited an infection sustained by a different HPV genotype at the follow-up visit compared to that identified at baseline.
DISCUSSION
Limited data are available regarding the distribution of oropharyngeal HPV types in HIV-positive subjects, and more specifically, in HIV-infected MSM. Knowledge of the distribution of specific HPV types in populations at high risk for HNSCC would be very useful for understanding the need for HPV prevention strategies. In the present study, a high diffusion of both HR and LR alpha-HPV infections was detected in oropharyngeal mucosa samples that were collected from HIV-infected MSM in an Italian population. Moreover, a broad range of the HPV genotypes identified belonged to groups 1, 2, and 3, according to IARC classification [13].
Previously, mucosal alpha-HPV genotypes were associated with infections that manifested in the genital tract [13]. In the present study, mucosal alpha-HPV genotypes were detected in the collected oropharyngeal mucosa samples, and these results suggest a possible sexual transmission as a primary route by which oropharyngeal HPV infections are spread in HIV-infected MSM.
The use of a DNA sequencing method was useful for identifying 22 different oral HPV genotypes. Despite the fact that sequencing does not allow detection of potential co-infections, this method was chosen for its ability in identifying all infecting types, including those not detected by commercially available kits and/or rare genotypes.
In the present study, two of the oropharyngeal swab samples contained uncommon alpha-HPV genotypes, namely HPV-102 and HPV-114. To our knowledge, this is the first report of these rare types being identified in the oropharyngeal tract, although cervical infections supported by these two genotypes have been described [Reference Miyashita15, Reference Ekström, Forslund and Dillner16]. This finding seems to strengthen the hypothesis of HPV transmission through oro-genital sexual contact.
The percentage of HR genotypes detected in the present cohort was higher than the percentage of LR genotypes (P < 0·05), with HPV-16 and HPV-33 being the most commonly identified HR genotypes. Of the LR genotypes, HPV-6 and HPV-32 were the most prevalent. Currently, the clinical significance of oropharyngeal HPV type-specific infections is unknown, although accumulating evidence supports a causal role for HPV-16 in a subset of head and neck cancers [Reference Ekström, Forslund and Dillner16]. Furthermore, other HR HPV genotypes have been associated with the development of different cancers [Reference Bouvard17]. Little is also known about the clinical relevance of LR HPV oropharyngeal infections.
The present study provides evidence that oropharyngeal HPV infections have a relevant epidemiological impact in HIV-positive MSM. Both the HPV prevalence (24·4%) and the cumulative incidence (24·1%) are substantial and are also consistent with the data reported in other studies conducted in Italy and in other countries [Reference Beachler6, Reference Mooij7, Reference Parisi18–Reference Ong20]. In Northern Italy, Parisi et al. [Reference Parisi18] reported a prevalence of oral HPV infection of around 20%, and Ong et al. found a prevalence of 19% in Australian HIV-positive MSM [Reference Ong20]. Moreover, the cumulative incidence (15·5%) of the HR infections detected in the present cohort is similar to the 6-month incidence rate (14·1%) reported by Mooij et al. in HIV-infected MSM in The Netherlands [Reference Mooij19]. Other studies have described lower rates of HR infections [Reference Beachler6, Reference Ong20], possibly due to a lack of suitable sampling and standardization of HPV testing for this anatomical site.
The results of previous studies have suggested that a substantial proportion of HPV oropharyngeal infections clear within 1 or 2 years, and this clearance period is modestly faster than that reported for anogenital infections [Reference Beachler6, Reference Parisi18, Reference Ong20]. This difference in clearance may be due to the characteristics of local mucosal immunity at these two sites. Only one persistent case of HPV-33 infection was detected during the follow-up period in the present study. Thus, it appears that most HPV oropharyngeal infections are transient and resolve over a short period of time.
However, the evolution of oropharyngeal HPV infections needs to be better investigated in future studies with a larger cohorts and with the use of genotyping methods that are able to identify potential HPV co-infections. A weakness of this study is that the sequencing method used may have underestimated type-specific prevalence and incidence. In this regard, at follow-up four subjects exhibited a HPV infection sustained by a type different from that detected at baseline. It is difficult to interpret this result as a new infection (incident infection), rather than a change in predominance between two or more infections simultaneously present in the same subject.
Recent studies have reported that severity of immunosuppression is one of the stronger risk factors for the acquisition of an oral HPV infection [Reference Denny4, Reference Beachler21]. Other risk factors include low CD4 cell counts and higher HIV viral loads. In the present study, the association between HPV infection and HIV status could not be evaluated due to the small number of subjects analysed, most of which were immune-reconstituted by highly active antiviral therapy.
In conclusion, the results of the present study may help to clarify the epidemiological impact of oropharyngeal HPV infections in HIV-infected MSM. Oropharyngeal HPV infection is common in Italian HIV-positive MSM, and these infections are associated with a broad spectrum of genotypes, mostly belonging to the HR clade. Of note, we found two rare alpha-types, namely HPV-102 and HPV-114, currently identified only in the genital mucosa. This is the first evidence of oropharyngeal infection associated with these types.
Future studies are needed to investigate the clinical impact and the natural history of infections related to HPV genotypes identified in the oropharynx. This will allow establishment of the necessary specific prevention strategies, both primary and secondary.
DECLARATION OF INTEREST
None.





