Biological materials have long been a source of both inspiration and envy to materials scientists. Wood, bone, nacre, silk, and muscle all show remarkable properties such as toughness, strength, and adaptability, while being produced from widely available feedstocks using methods that, by definition, are sustainable and environmentally friendly.
But while there have been many efforts to mimic the structures and properties of these natural materials—the field of biomimetic materials is large—we haven’t yet tried to reap many of the benefits of their means of production. Arguably that’s where nature’s real superiority lies. The mechanical virtues of silk, for example, arise from the precise control that organisms such as spiders and silkworms have over not only the complex chemical compositions of these protein-based polymers but also factors such as fiber orientation and hydration during the spinning process, which introduce a hierarchy of structure into the final product (see Figure 1). By tweaking the composition anigurd processing, these organisms can produce different types of silk fiber “engineered” for different roles. It’s not easy to see how we can achieve comparable materials properties—or tailor new ones from the same raw ingredients—without some access to the means of production.

Figure 1. Silk hierarchical structure, from microns, to nanometers, to molecule-scale.
This is the next frontier for making use of biological know-how in materials synthesis and discovery. “I believe that the future of manufacturing will be, in large part, driven by biology,” said Patrick Boyle, head of design at Gingko Bioworks, Inc. in Boston, one of the first companies to try to develop highly engineered organisms—so-called synthetic biology—for industrial-scale processing of chemicals and materials.
The possibilities are vast but so are the challenges, because using living organisms to make bespoke materials takes materials science far beyond its traditional skill set and demands that engineers sit down with molecular and cell biologists and geneticists, among others, to figure out how to put nature to work. “Materials science and biology speak different languages,” said synthetic biologist Christopher Voigt of the Massachusetts Institute of Technology, “and they are good at using different types of material.” The question is how to make them work together.
Nature as an engineering challenge
Engineering living organisms, particularly microbes such as bacteria, to make useful products has a long history, having been one of the aims of biotechnology since its early days. Human insulin for treating diabetes was first expressed in genetically engineered Escherichia coli (E. coli) bacteria in 1979, and was marketed from 1982 as the first licensed drug made by the then-new recombinant DNA technology. Today, hundreds of pharmaceutical products are made by fermentation of engineered microorganisms, and insulin is also manufactured in yeast.
Such products generally require little more than the transfer of a gene encoding the target protein from one organism to another. This cross-species exchange of genes can be dramatic, as in the case of goats given a spider silk gene so that they express the protein in their milk, a process developed in 2010. All the same, the degree of genetic manipulation involved is rather small: You take a DNA “instruction” from one genome and plant it in another.
But we can get much more ambitious than that. Structural proteins such as silk can be rather wonderful, but they represent only a tiny segment of the materials universe. Some useful natural substances are the result of a complex series of biochemical steps; this is true of many natural products with valuable pharmacological action (e.g., alkaloid compounds produced by plants as defensive agents). These molecules often have complicated structures assembled step by step by a suite of enzymes—the sequence and timing of the steps has to be just so.
Transferring such a multistep manufacturing process from one organism to another is far less easy. Traditionally, it goes by the name of metabolic engineering: recreating or adapting an entire metabolic pathway for a specific metabolite. Such efforts are also now commonly ushered under the broad umbrella of synthetic biology, a term that refers to a philosophy as much as to a scientific discipline. Synthetic biology approaches living organisms much as engineers approach machines—as things that can be taken apart and put back together, rationally designed for particular purposes, and made to work on an industrial scale (see Figure 2).
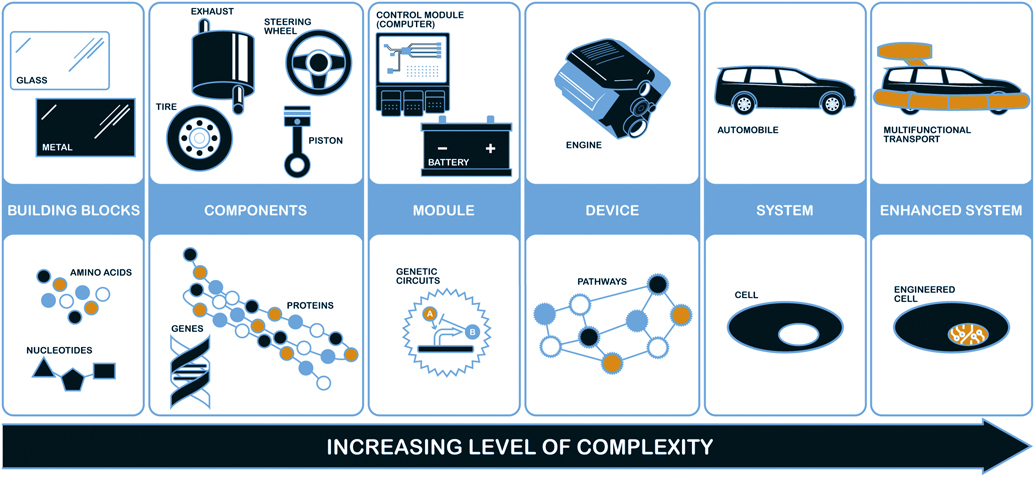
Figure 2. Parallels between regular engineering (top) and synthetic biology (bottom) extend from basic ingredients on the left to advance function systems on the right. Credit: Richard Eldridge, US Air Force Research Laboratory.
So far, much of the focus of synthetic biology has been on the production of useful chemicals: drugs and fuels, for example, both of which can be derived from compounds produced naturally. Others see synthetic biology as a potential source of organisms that perform useful functions, acting as sensors that signal the presence of substances such as toxins or disease markers in the body or the environment. Some organisms engineered by synthetic biologists exhibit gloriously quirky properties, such as light sensitivity that turns them into “living cameras,” or the ability to emit periodic flashes of light (see Figure 3).
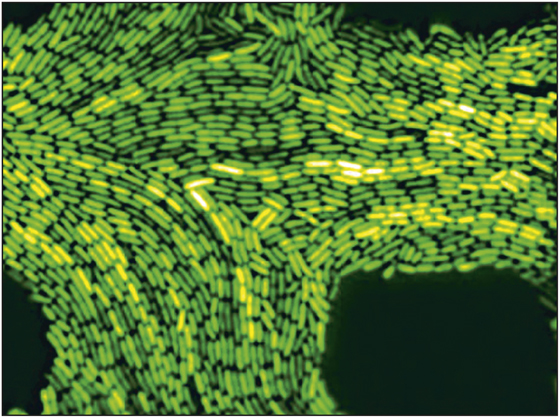
Figure 3. Green fluorescent protein causes the E. coli to glow when the cells’ clock is activated. Credit: UC San Diego.
All of these objectives overlap with some of the functions associated with materials engineering. But only recently has materials production started to emerge as an explicit objective of synthetic biology. It’s an especially challenging goal but also a particularly exciting one. For synthetic biology might supply a means not just to make the basic chemical ingredients of new materials, but to give them the kind of sophisticated, hierarchical, and nanoscale structures that hold the key to the superior performance of many biomaterials. In other words, it’s here that we might start to find the first attempts to leverage the extraordinary organizational, architectural power of living nature to build structures from the bottom up.
The birth of synthetic biology
Synthetic biology is arguably a new name for an old idea. In his 1912 book The Mechanistic Conception of Life, German American biologist Jacques Loeb proposed that living organisms be regarded from an engineering perspective—not just as entities “engineered” by evolution, but as devices amenable to human intervention, manipulation, and alteration. “The idea is now hovering before me,” he wrote in 1890, “that man himself can act as creator even in living nature, forming it eventually according to his will.”
That was a vision born partly out of sheer naivety, though. Where life’s vitality came from had long been a deep mystery to scientists, but only in the decades following Loeb’s proposal did biologists start to see that the origins sprung from the capacity of living matter to organize itself at microscopic scales. And that organization soon looked overwhelmingly complicated.
In some ways it still does. Hopes that our understanding of the mechanisms of genetic inheritance and control—the transmission, mutation, and translation of information encoded in DNA—will lead us to comprehend cells and organisms in terms of their molecular machinery have given way to an appreciation of the baffling complexity of the genome. Life proceeds from the orchestration of gene activity, responding to and modified by its environment, in ways that seem bewildering and which have outstripped our simplistic models derived from computer theory and cybernetics.
All the same, we evidently can intervene in genetics in ways that are sometimes predictable. We understand a great deal about how genes tend to work in networks, regulating one another’s activity to direct the fate of the cell. This analysis of genomes in terms of functional networks involving feedback, switching, and amplification is one that sounds familiar to engineers, and the hopes and dreams of synthetic biology ride on it.
The essence of that mind-set is nicely captured by historian of science Sophia Roosth of Harvard University in her recent book Synthetic, a sociological survey of the emergence of synthetic biology. She describes how when Drew Endy of the Massachusetts Institute of Technology (MIT) developed a computational model of the genetics of the bacterial virus T7 as a doctoral student in the late 1990s, he found that it dismally failed to predict how the virus would respond to perturbations in the environment. But whereas most researchers would have gone back to the drawing board with their model, Endy decided that it was the virus that needed to be taken there. The model was fine; the problem was that the virus didn’t conform to it. It needed debugging.
In other words, the aim of synthetic biology is fundamentally different, even opposite, to that of biology. It doesn’t take nature as an object of study, trying to figure out why it does the things it does. Rather, you start with an objective, and ask how biology can be engineered—some might say “improved”—to do what you want it to.
MIT has always been one of the hot spots of synthetic biology, to the extent that some feel it has imposed its philosophy unduly on the whole field. In the early 2000s, Endy joined forces with veteran MIT engineer and computer scientist Tom Knight to found the Synthetic Biology Working Group. Their aim was to make synthetic biology a systematic engineering discipline. They proposed to create a registry of standardized parts for engineering organisms such as bacteria and viruses: a set of “gene circuits” with well-defined functions designed to strict specifications so that they could be interfaced on a plug’n’play basis, just like the standard components of electronic circuits. The MIT group also initiated an annual event called the iGEM (International Genetically Engineered Machine) competition in which student teams use biological parts to create innovative functions in living cells. This event arose out of an MIT course in 2003 in which students made bacterial cells that “blinked” on and off with light. Knight was also the driving force behind Gingko Bioworks, a company spun out to turn some of the advances made in synthetic biology in academia into real manufacturing processes that work on a large scale.
Genes for polymers
While this mind-set of engineering and computer science underpins much of synthetic biology, the discipline has other flavors too. Some of it looks like an elaboration of more conventional genetic and metabolic engineering; it’s something of a moot point where genetic engineering using well-established recombinant DNA technology to insert new genes into non-native genomes turns into synthetic biology. “There is a long history of applying recombinant protein techniques to produce protein-based materials,” said Chao Zhong, a synthetic biologist at ShanghaiTech University in China.
Some of that early work involved energy-storage polymers called polyhydroxyalkanoates (PHAs) made naturally by some bacteria, which could be processed into biodegradable plastics.Reference Doi1 For example, genes from the PHA-producing bacterium Alcaligenes eutrophus were transferred into the more readily fermented “workhorse” bacteria E. coli or even into oil-seed rape plants from which the polymers could be extracted in oil.
“Such ‘old’ molecular biology tools can be coupled with rational design principles to produce bioinspired molecular materials that may outperform existing or natural biomaterials,” said Zhong. For example, while working at MIT, Zhong and his co-workers genetically engineered E. coli to produce proteins that merged a fiber-forming polypeptide called CsgA, made naturally by the bacterium, with a highly adhesive protein made by mussels, creating a material that self-assembles into a strong underwater adhesive.Reference Zhong, Gurry, Cheng, Downey, Deng, Stultz and Lu2
This was basically a matter of abutting two separate genes end to end. But efforts to engineer organisms to make new substances can get more complicated. When you need several genes added to make a particular product, you have to start worrying about the genetic networks involved in switching them on or off at the right times, and that’s when it all starts to look more like circuit design.
The flagship example of this kind of thing is the work conducted at the University of California, Berkeley, by bioengineer Jay Keasling and his co-workers. For several years, they have attempted to equip microbes such as bacteria and yeast with the genetic tools needed to produce artemisinin, a natural product found in a species of the East Asian plant, sweet wormwood (Artemisia annua), which has antimalarial properties.Reference Ro, Paradise, Ouellet, Fisher, Newman, Ndungu, Ho, Eachus, Ham, Kirby, Chang, Withers, Shiba, Sarpong and Keasling3,Reference Keasling4 As with many natural products, the natural sources of this valuable drug are scarce: It is made only in tiny amounts in the leaves of the sweet wormwood shrub. So a way to produce it by biotechnological fermentation could be of great value.
Making organisms that produce rare and precious substances is at the core of much of the work at Gingko, said Boyle. Since the early 2010s, when the company had only a few dozen staff, it has expanded to hundreds, and it works together with clients to find synthetic-biology solutions to manufacturing problems such that they can be carried out in a scalable fashion. Boyle said that the factory (“foundry”) lab space now covers 20,000 square feet and is built around automated procedures.
Gingko’s early work was geared toward fine chemicals: making microbes that produce flavorings and fragrances. But materials, particularly polymers and their molecular precursors, are now very much on their agenda. Typically, said Boyle, synthetic polymers have been geared to what you can make from oil. “But if you look at the natural world, there’s a whole host of different structures that look nothing like what you can make from petrochemicals—not just proteins but carbohydrate polymers and novel metamaterials in which specific structures are arranged.” An example of the latter is the periodic porous lattice of some butterfly wing scales, made from the polysaccharide chitin, in which the regular array of voids makes the material a photonic crystal that diffracts light and creates structural color.
Gingko is currently working with the US Air Force Research Laboratory at the Wright-Patterson Air Force Base in Ohio to develop a production process for a polymer called Nvjp-1, a remarkably strong protein found in the jaws of a marine sandworm (see Figure 4). “Nvjp-1 has some interesting and reversible mechanical properties that we would like to explore for composite structures,” said Rajesh Naik of the Air Force team. “We were able to recombinantly express and purify the protein from bacteria, and Gingko has been scaling up the expression to create composite structures for real-world application and testing.”
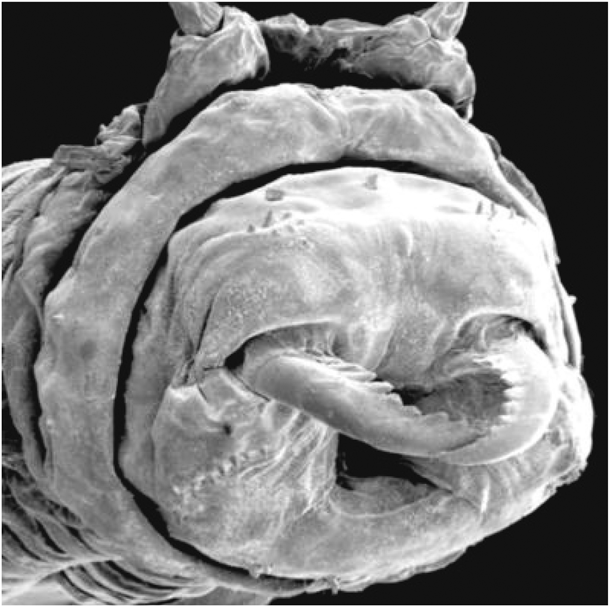
Figure 4. Researchers are reporting that the protein composition of the fang-like jaws of Nereis virens, a common marine worm, could be used in construction and aerospace. Image courtesy of Chris Broomell; https://phys.org/news/2008-07-marine-worm-jaws-cutting-edge-aerospace.html.
One ultimate goal here might be a kind of digital synthetic biology in which a peptide-encoding gene is designed from scratch on a computer, the corresponding DNA is synthesized chemically, and the gene is inserted into the genome of a cell to make the material. That vision, however, will depend on developing a better understanding of how a protein’s structure and physical properties are determined by the primary amino-acid sequence—a question that depends, in part, on how protein chains fold and are hydrated, issues that are still generally rather far from being routinely predictable.
Synthetic biology needn’t yield a finished material; it could simply be a good way of getting the raw ingredients. Another material commodity for which biological production is being developed at Gingko, in conjunction with the San Diego-based company Genomatica, Inc., is a monomer called 1,4-butanediol (BD), used to make elastomers such as polyurethanes and Spandex. In processes like this, a key question for commercial viability is whether the product can be created from a sufficiently cheap and abundant feedstock; that’s to say, whether you can persuade your organisms to consume and transform the right stuff.
Finding a metabolic pathway to the target product is also a question of identifying enzymes that will do the required chemistry. Those could be anywhere: in plants, insects, bacteria, fungi. Finding them is often a massive exercise in bioprospecting, perhaps augmented by some retooling of an enzyme to give the right product specificity. “Typically we look at thousands of different natural enzymes to find the best candidates,” said Boyle. Once they are found, researchers will order synthetic DNA encoding of that molecule. (Boyle said that Gingko is the single largest consumer of synthetic DNA in the world.)
Which organism do you then put it into? Boyle said that the company spends a lot of time looking for good “chassis” strains: organisms that work well as standard vehicles for gene expression. They work routinely with around five different types of bacteria, as well as yeast and fungi.
It seems likely that synthetic biologists will continue to use familiar, existing organisms such as E. coli and the yeast Saccharomyces cerevisiae as their standard chassis for the time being. But some synthetic biologists suspect it might ultimately be best to design the host organisms from scratch, stripping them down for optimal efficiency to only the genes needed for the job. In 2010, biotech entrepreneur Craig Venter and his colleagues at the J. Craig Venter Institute in La Jolla, Calif., “redesigned” the genome of the microbe Mycoplasma mycoides, built the DNA chemically from scratch, and transplanted it into cells of a closely related microbe to “reboot” it as an allegedly “synthetic organism.”Reference Gibson, Glass, Lartique, Noskov, Chuang, Algire, Benders, Montague, Ma, Moodie, Merryman, Vashee, Krishnakumar, Assad-Garcia, Pfannkoch, Denisova, Young, Qi, Segall-Shapiro, Calvey, Parmar, Hutchison, Smith and Venter5 In 2016, Venter’s team reported that they had streamlined the M. mycoides genome, removing anything deemed unnecessary, to make a “minimal organism” with only 473 genes (the functions of many of which remain unknown).Reference Hutchinson, Chuang, Noskov, Garcia, Deerinck, Ellisman, Gill, Kannan, Karas, Ma, Pelletier, Qi, Richter, Strychalski, Sun, Suzuki, Tsvetanova, Wise, Smith, Glass, Merryman, Gibson and Venter6 Not only might such efforts provide a simple, versatile chassis for carrying designed gene circuits, but they could also offer a way to add biosafety measures into organisms engineered for synthetic biology: You could design them, for example, only to be able to metabolize substances not found in the natural environment.
Expanding nature’s repertoire
Given that nature supplies a polymer-making factory of such remarkable precision and dexterity—the ribosome on which sequences from DNA are converted into sequences of amino acids in proteins—it seems a shame that it operates only on a limited palette of monomers. True, the 20 amino acids found in proteins can be permuted into a literally astronomical variety of sequences, of which even nearly 4 billion years of evolution could have explored only a small fraction. What’s more, proteins already show a wide range of materials properties, from the tough disorderly collagen matrix of composites such as bone to the optically transparent cystallin of the eye’s lens and cornea. All the same, an ability to add new, non-natural residues to proteins could broaden the functionality of these polymers beyond what nature can supply, giving them useful magnetic or electronic properties, for instance. “The extraordinary synthetic capability of the protein biosynthesis system has driven extensive efforts to harness it for societal needs in areas as diverse as energy, materials, and medicine,” said chemical engineer and synthetic biologist Michael Jewett of Northwestern University in Evanston, Ill., who is expanding the range of amino acids that this system can accept and incorporate into proteins.
Such semisynthetic materials might have countless uses, he said, for making both fine chemicals and new materials. “Expanding nature’s repertoire of ribosomal monomers could yield new classes of enzymes, therapeutics, materials, and chemicals with diverse genetically encoded chemistry. Sequence-defined polypeptides made of protease-resistant monomers, which would not be readily digested by cells, could lead to antimicrobial drugs that combat rising antibacterial resistance, and one could imagine making functional self-assembling nanoparticles that could be used for drug delivery,” said Jewett.
The basic idea goes back at least two decades. In the 1990s, David Tirrell of the University of Massachusetts Amherst force-fed bacteria on a non-natural fluorinated derivative of the amino acid phenylalanine to encourage the emergence of strains that would accept only this version for incorporation into proteins.Reference Yoshikawa, Fournier, Mason and Tirrell7 Tirrell and colleagues speculated that if these fluorinated groups were suitably placed, the polypeptide chain might fold into a kind of “Teflon-coated,” nonstick structure. Tirrell also attempted to create proteins that would fold into films with non-natural amino acids at their surfaces bearing monomer units that might be cross-linked into conducting polymers.
Synthetic biology could make it possible to create such materials by redesigning the molecular machinery that puts it together, rather than by forced evolution. Harnessing this apparatus could offer unprecedented control over the chemical structure of the products. “Biology enables more precise control of the final product and sustainable synthesis,” said Jewett. “Long sequence-defined polymers can be made that are not possible using traditional chemical approaches.”
But getting this machinery to work with non-natural amino acids is a tall order, he added—not least because it also needs to keep making the regular proteins on which the viability of the cell depends. “Because a functioning ribosome is critically necessary for life, cell viability restricts the mutations that can be made to it, said Jewett. “In practice, these constraints have made the natural ribosome essentially un-evolvable. And until recently, there has not been a generalizable method for modifying the ribosome’s active site to work with substrates beyond those found in nature.”
Proteins aren’t the only useful biopolymers. There has been relatively little work on polysaccharides and glycoproteins—both immensely versatile and valuable in biology—because their compositions can be very complicated, and their production process and structure–property relationships are still not well understood. But another biopolymer that is increasingly being appreciated for its materials virtues is DNA (see the December 2017 issue of MRS Bulletin), which researchers can design both to make self-assembling nanostructuresReference Rothemund8 and for high-density information storage.Reference Erlich and Zielinski9
Here too, using non-natural building blocks (nucleotides) could expand the possibilities for materials science. “DNA’s functionality is really limited, and it is difficult to control it and attach things to it in any ordered way,” said Floyd Romesberg of The Scripps Research Institute in La Jolla, Calif. “So you cannot really take advantage of the unprecedented compositional and spatial control it offers, because you cannot specifically attach things to the nucleotides.” His group uses a non-natural base pair added to DNA at defined positions as a “hook.” They have evolved a new kind of polymerase—the enzyme that strings DNA nucleotide bases together—that will tolerate non-natural bases and have shown that they can add functionalized bases with azido, chloro, and amino substituents.Reference Chen and Romesberg10 This enabled Romesberg and colleagues to create a DNA-based polymeric material, a hydrogel. Romesberg’s work is behind a new startup called Synthorx, which aims to use the non-natural base pair to develop new therapeutic proteins.
The great attraction of using DNA and proteins as materials, despite their limitations, is that they are evolvable, so one can imagine using directed evolution in vitro to get new, non-natural functions. “Imagine you want a ‘wire’ that conducts electricity,” said Romesberg. “You could set up a selection process where if a piece of DNA does not conduct, it would be degraded; for example, if a high-energy electron is efficiently transferred to some other material, the DNA remains intact, but if it is not, it cleaves the DNA backbone. You could subject them to these conditions and then amplify [using the standard polymerase chain reaction] the ones that survive. The ability to evolve biological materials makes them rather different.”
What’s more, they can be produced in vivo. While there is interest in using synthetic DNA nanostructures to intervene in and interrogate living cells,Reference Chen, Groves, Muscat and Seelig11 potentially one might close that circle by exploiting the ability of cells to make nucleic acids. There are already some steps in this direction. Timothy Lu at MIT, for example, is working with Fahim Farzadfard to program E. coli to make single-stranded DNA molecules in response to specific signals from gene regulatory circuits, thereby creating a permanent material memory of transient events in the cell.Reference Farzadfard and Lu12 Such an in vivo DNA memory can be made more sophisticated by harnessing the precision DNA-editing machinery of the CRISPR-Cas system to write information stably into a cell’s genome. In this way, Harris Wang at Columbia University in New York and colleagues made E. coli that could record the ephemeral availability of different metabolites in their environment over time.Reference Sheth, Yim, Wu and Wang13
Managing many scales
One of the most striking features that sets apart some natural materials, such as structural proteins, from synthetic ones is that they exhibit structures over a range of length scales: so-called hierarchical structure.Reference Reznikov, Bilton, Lari, Stevens and Kröger14 Whereas polymer scientists can, for example, control the degree of bulk crystallinity in a synthetic hydrocarbon polymer by dictating the shape of its molecular chains and their side groups, the complex yet precisely specified amino-acid sequence of silks may dictate a sophisticated microstructure in which nanocrystalline regions are interspersed among a more disorderly matrix.
“Decades of fundamental research on biomaterials have provided unique insight into their constituents and hierarchical organization from atomic to mesoscales, and their resulting exceptional functional properties,” said Jewett. He explained that while most biomaterials employ proteins as key building blocks with specific amino-acid sequences, those primary structures might be elaborated into coils, sheets, and other folds (the secondary structure) and further aggregated or cross-linked to achieve specific functions such as energy storage, extensibility, strength, hardness, and toughness.
Could materials made using synthetic biology have access to this capacity for structural control over many scales? Jewett hopes so. “We envision a future [of] materials synthesis that arises from hierarchical assembly of polymers,” he said, “first by making uniform building blocks with precise functionality, then assembling them into substructures, and, finally, locking them together into polymeric megamolecules with unique, complex, and highly useful properties, including biomechanical and catalytic properties as well as biophysical selectivity arising from the control of surface characteristics.”
The key to achieving such hierarchy, said Voigt, is twofold. One needs not only to manipulate the synthetic pathways that an organism uses, as in the case of metabolic engineering of artemisinin, but also to control the timing of gene activation, switching them on and off in the correct sequence. That, after all, is how cells turn into complex hierarchical structures like us, by an elaborate unfolding in time that patterns initially identical cells into different tissues.
Alongside their structural hierarchy, many biomaterials achieve their enviable characteristics via a composite structure—through an intimate, carefully controlled mixture and arrangement of a variety of substances, both organic and inorganic. That’s seen, for example, in the combination of phosphate crystallites and collagen matrix in bone, or the layers of crystalline calcium carbonate sandwiched between soft protein sheets in nacre shell (see Figure 5).

Figure 5. A close-up of the spirals of a chambered nautilus made from the layered composite material nacre (Nautilus pompilius).
This combination of inorganic and organic has been a challenge to achieve in conventional biomimetic materials engineering. Traditional biopolymers used for materials such as cotton and silk are all carbon-based, said Voigt, but “biology makes so much more than that.” He points to the delicate yet strong latticed frameworks of diatoms, aquatic microorganisms made from silica (see Figure 6). They’re manufactured in a dirty, noisy environment, he said, yet “with a precision, you can’t get in a clean room,” the typical environment needed for high-precision, ultrapure manufacture of microelectronics.

Figure 6. Diatoms under a microscope.
Living organisms exploit such organic–inorganic composites not just for their mechanical properties but for other functions too. Some bacteria, for example, make magnetosomes: small particles of iron oxide organized into chains that serve as magnetic sensors, allowing the microorganisms to detect and respond to magnetic fields. These particles “have a precise, genetically controlled shape and size,” said Boyle, and they offer tantalizing evidence of how living systems might be harnessed to generate structures more commonly associated with precision engineering—in electronic circuits, memories and sensors, and beyond. “Engineered cells could be coupled with inorganic semiconductor materials to form new biotic–abiotic interfaces to convert solar energy into chemical energy through artificial photosynthesis,” suggested Zhong.
Boyle suspects that nature might contain many proteins that could be exploited for interfacing with inorganics. Copper-accumulating organisms are already used, for example, for recovering that metal in mining. “Maybe we can look for proteins that coordinate interesting metals in nature, and then look for ways of structuring them with protein-based scaffolds,” said Boyle. The point here could be simply to use the organisms as a kind of workforce for assembly, and then get rid of them. “The biology doesn’t need to stick around,” he added.
Creating organisms that make new materials could be immensely valuable as a new means of achieving molecular precision from processes that are renewable almost by definition. But biology promises more, because one of the key attributes of living systems is that they don’t just make stuff but do stuff: They are adaptive and responsive to their surroundings. This active functionality is what appeals to synthetic biologists. In the early days of the field, this was displayed by engineered microbes that could oscillate and emit light periodicallyReference Elowitz and Leibler15 or with gene circuits that could be switched on and off like toggle switches in electronic circuits.Reference Gardner, Cantor and Collins16,Reference Hasty, McMillen and Collins17 One of the most eye-catching successes was the development by Voigt, then at the University of California, San Francisco, and co-workers of a strain of E. coli bacteria that could act as a kind of living photographic film. They respond to light by switching off a gene that otherwise produces an enzyme catalyzing the formation of a black precipitate in the growth gel medium. Voigt’s team has since refined the method to produce “bacterial images” in other ways at high resolution (see Figure 7).Reference Levskaya, Chevalier, Tabor, Simpson, Lavery, Levy, Davidson, Scouras, Ellington, Marcotte and Voigt18

Figure 7. Light is used to induce cells to produce a biomaterial, specifically a functionalized bacterial amyloid fiber. With optogenetics, remarkable resolution and control of this biomaterial are achieved. The top pattern is the light mask (an extrapolation of a da Vinci sketch) that is shone on a liquid culture of engineered E. coli, and the bottom is the resulting biomaterial produced by the cells and then stained with crystal violet dye on a petri dish. Credit: Christopher Voigt and Felix Moser, Massachusetts Institute of Technology.
The possibilities for sensing technologies are obvious: These are potentially very sensitive detectors that can actively explore their environment, like miniature robots. This is where synthetic biology could take materials science into truly new territory. “One genuinely new area that synthetic biology can bring into materials science is to develop so-called living functional materials,” said Zhong.Reference Chen, Zhong and Lu19 Such materials in nature include biofilms, shells, and skeletal tissues—multifunctional, multiscale, environmentally responsive assemblies of living and nonliving components. “They grow, self-repair, and adapt to the environment, and could be used to produce smart materials that dynamically respond to their surroundings,” said Zhong.
There are already examples of living functional materials. Lu and colleagues at MIT have engineered E. coli so that they will make a fibril-forming protein called curli only in response to specific chemical signals from the environment.Reference Chen, Deng, Billings, Seker, Lu, Citorik, Zakeri and Lu20 The fibrils self-assemble over a range of length scales determined by the proteins’ primary sequence, creating patterned biofilms. The researchers incorporated tags into the protein chains that can latch onto inorganic nanoparticles (e.g., gold or cadmium sulfide quantum dots), and thereby use the fibrils as self-assembling templates with a tunable nanostructure for patterning the particles with precision in a process that is switchable and controllable.
This system, in which curli fibrils are formed by a well-defined gene circuit, is proving rather versatile for making responsive nanopatterned materials. Lingchong You and his co-workers at Duke University have used it to assemble pressure sensors.Reference Cao, Feng, Ryser, Zhu, Herschlag, Cao, Marusak, Zauscher and You21 They programmed their curli-making bacteria by engineering a gene circuit to self-assemble into a three-dimensional shape on a porous membrane that dictated the structure of the curli biofilm and the inorganic components it templated. They used inkjet printing to create an array of bacterial colonies on the membrane, each of which grew into a hemispherical shape covered by a dome of curli fibrils to which gold nanoparticles could be attached. By wiring up these gold-coated structures, they made pressure sensors in which pressure applied to the domes could flatten them and bring adjacent ones in contact to generate a conductive pathway. The touch-sensitivity of the structure depended on the precise shape of the domes, which itself may depend on the membrane properties.
Recently, Zhong has programmed curli-producing E. coli to do so only in response to blue light, enabling the researchers to use light to write into a biofilm pattern to which fluorescent nanoparticles can be attached with a spatial resolution of 100 micrometers.Reference Wang22 “I believe there is a great future in this area to leverage synthetic biology tools to reprogram and control cell behavior so that material building blocks could be produced and secreted in a spatiotemporally controlled manner,” said Zhong.
Scaling up
In theory—and in the lab—the possibilities seem endless. In practice, however, is another matter. The challenges of scaling up lab-based discovery into commercial manufacturing are daunting, but that’s what Gingko hopes to crack. “Aside from obvious targets of synthetic efficiency,” said Boyle, “a key aim is to bed down such manufacturing processes among the existing industrial ecology, for example, to make sure that the feedstock of a biological strategy is cheaply and abundantly available.” With canny planning, different processing tasks could support one another, the byproducts of one metabolic pathway being useful for another. After all, he said, the petrochemicals industry has evolved so that no fractionate of crude oil goes to waste. Creating such an economical, sustainable web of processes for mass production synthetic biology might, “cause a rethink of the entire supply chain.”
Boyle draws an analogy with the founding of industrial synthetic chemistry in the 19th century, in which the byproduct of extracting gas from coal, called coal tar, which was thought to be a useless waste residue, became the crucial feedstock for the production of synthetic dyes. This led dye companies to diversify into pharmaceuticals and other fine chemicals and to establish R&D departments to develop basic science. Brian Yeh and Wendell Lim of the University of California, San Francisco, have pointed out how development in 19th-century organic chemistry made it turn toward synthesis, which then led to the fundamental understanding of structure and reactivity. Perhaps synthesis in biology will have the same effect.Reference Yeh and Lim23
This comparison teaches another important lesson. The discovery of coal-tar dyes was a serendipitous accident (arising from an attempt to make the antimalarial quinine), taking applied synthetic chemistry in a direction no one had anticipated. The same might be true of synthetic biology as a manufacturing technology: no one has yet found the “killer app.” “Where we’re going may be quite different from where it seems today,” said Boyle. Just like evolution, really.