1. Introduction
Bacterial blight (BB) of rice, caused by Xanthomonas oryzae pv oryzae (Xoo) (Ishiyama, Dye), is found worldwide and is particularly endemic to much of Asia (Mew et al., Reference Mew, Alvamz, Leach and Swings1993; Nino-Liu et al., Reference Nino-Liu, Ronald and Bogdanove2006). At seedling stage (Kresek), the disease may cause complete wilting or death of effective tillers, whereas at maximum tillering stage, it results in blighting of leaves. Losses due to its outbreak can reach up to 50% depending on stage, weather, location and variety (Reddy et al., Reference Reddy, Mackenzie, Rouse and Rao1979; Mew et al., Reference Mew, Alvamz, Leach and Swings1993). The Xoo pathogen is highly variable and over 30 races of it have been reported worldwide (Nino-Liu et al., Reference Nino-Liu, Ronald and Bogdanove2006; Gonzalez et al., Reference Gonzalez, Szurek, Manceau, Mathieu, Séré and Verdier2007). Because of variability in virulence, breeding of resistant cultivars always confronts difficulties in terms of durability of resistance. Genome structure of Xoo revealed the presence of a large number of effector (avr) genes and insertion sequences (Ochiai et al., Reference Ochiai, Inoue, Takeya, Sasaki and Kaku2005; Lee et al., Reference Lee, Park, Park, Kang, Kim, Song, Park, Yoon, Hahn, Koo, Lee, Kim, Park, Yoon, Kim, Jung, Koh, Seo and Go2005), which may be playing a major role in generating high degree of genetic diversity and race differentiation (Ochiai et al., Reference Ochiai, Inoue, Takeya, Sasaki and Kaku2005). In Punjab State of India, for example, two pathotypes were reported in the 1980s, but this number has now grown to seven (Lore & Raina, Reference Lore and Raina2005; J. S. Lore, personal communication), despite the fact that BB-susceptible cultivars occupied a large area under cultivation during this period. Chemical control of BB is not possible because the effective and economical chemical control measures are not available (Gnanamanickam et al., Reference Gnanamanickam, Priyadarisini, Narayanan, Vasudevan and Kavitha1999); therefore, the preferred strategy for BB management has been through development and deployment of resistant cultivars (Khush, Reference Khush2001).
Currently, more than two dozen genes conferring resistance on BB have been identified and these are designated Xa1 to Xa29(t) (Nino-Liu et al., Reference Nino-Liu, Ronald and Bogdanove2006). A majority of these are from cultivated rice, except Xa21, Xa23, Xa27 and Xa29, which have been transferred from Oryza longistaminata, Oryza rufipogon, Oryza minuta and Oryza officinalis, respectively (Khush et al., Reference Khush, Bacalangco and Ogawa1990; Zhang et al., Reference Zhang, Lin, Zhao, Wang, Yang, Zhao, Li, Chen and Zhu1998; Gu et al., Reference Gu, Tian, Yang, Wu, Sreekala, Wang and Yin2003; Tan et al., Reference Tan, Ren, Weng, Shi, Zhu and He2004). Not all the known genes are effective, but deployment of a few genes has been highly successful. Xa4, for example, has been used most widely in the tropics and in many parts of China (Sun et al., Reference Sun, Yang, Wang and Zhang2003; Mew et al., Reference Mew, Leung, Savary, Vera and Leach2004; Vikal et al., Reference Vikal, Goel, Singh and Singh2004), whereas Xa1 and Xa3 are widely used in Japan and Korea (Mew et al., Reference Mew, Leung, Savary, Vera and Leach2004). In the North Western Plains of India, except for xa13, none of the known Xa genes are effective singly against all the prevalent Xoo pathotypes. Also appearance of new pathogen virulences warrants expanding the gene pool of effective and potentially durable genes (Mew et al., Reference Mew, Leung, Savary, Vera and Leach2004). Most of the known BB resistance genes have been tagged with DNA markers (He et al., Reference He, Li, Zhu, Tan, Zhang and Lin2006) and six, viz. Xa1, xa5, xa13, Xa21, Xa26 and Xa27, have been cloned (Dai et al., Reference Dai, Liu, Xiao and Wang2007). Keeping in view the evolutionary potential of Xoo, identification of new genes or pyramiding of two or more genes is required for ensuring control of the disease. Here, we report identification and mapping of a new dominant BB resistance gene from Oryza nivara and its transfer to cultivated rice, Oryza sativa.
2. Materials and methods
(i) Plant material and population development
O. nivara acc. IRGC 81825 (hereafter referred to as acc. 81825) procured from IRRI (International Rice Research Institute; Los Baños, Laguna, Philippines) showed a high level of resistance to all the seven Xoo pathotypes tested (Vikal et al., Reference Vikal, Das, Patra, Goel, Sidhu and Singh2007). This accession was crossed as male to O. sativa cv PR114. The F1 plants were self-pollinated and backcrossed to PR114 to generate F2 and BC1F1 progeny. The resistant BC1F1 plants were again backcrossed to PR114 to generate BC2F1 that were further self-pollinated to generate BC2F2 generation. BB-resistant BC2F1 plants were again crossed to the recurrent parent PR114 to generate BC3F1 progeny. Individual BC2F2 plants were again selfed to generate BC2F3 progeny. The F2, BC3F1, BC2F2 and BC2F3 progenies generated were used for studying inheritance, mapping and stability of the resistance gene introgressed from O. nivara.
(ii) Screening with Xoo
The O. nivara acc. 81825 was resistant to all the seven Xoo pathotypes prevalent in Punjab (Vikal et al., Reference Vikal, Das, Patra, Goel, Sidhu and Singh2007) and PR114 was resistant to five but susceptible to two. Pathotype VII, which was virulent on PR114 but avirulent on O. nivara acc. 81825, was used for screening the segregating progenies. The Xoo pathotype VII was virulent on Xa1, Xa2, Xa3, Xa4, xa5, Xa7, xa8, Xa10, Xa11, Xa12, Xa14, Xa16 and DV85 (xa5+Xa7) but avirulent on xa13 and Xa21 (Table 1). The cultures, originally isolated from a single colony, were streaked on Walkimoto medium (10 g sucrose, 5 g peptone and 20 g agar; total volume made to 1000 ml using distilled water) slants in test tubes under aseptic conditions. After streaking, the cultures were incubated for 72 h at 28°C. The bacterial growth was scrapped and homogenized by vigorous shaking. The concentration of bacterium was standardized to a density of 108–109 cells per ml and used for inoculations. The BC3F1 and BC2F2 progenies were screened during 2006 and BC2F3 during 2007. Individual plants were inoculated at maximum tillering stage following clip inoculation (Kauffman et al., Reference Kauffman, Reddy, Hsieh and Merca1973). The disease severity was recorded 14–16 days after the inoculation. In this, lesion length in each plant was measured on five leaves and averages worked out. Plants having lesion length up to 5·0 cm were classified as resistant and the ones with more than 5·0 cm were classified as susceptible. This was based on the alley formed while classifying plants as resistant or susceptible. Chi-square test, after Yates correction (χC2), was used to test goodness of fit for ascertaining the number of genes governing the BB resistance. In addition to inoculations at maximum tillering stage, both O. nivara acc 81825 and PR114 were inoculated with pathotype VII at seedling stage (1-month-old seedlings) as well using the same procedure as described above.
Table 1. Avirulence/virulence (effective/ineffective) of Xoo pathotypes prevalent in Punjab (India) against known BB resistance genes

(iii) Marker analysis
DNA from parents and the segregating generations was isolated from field-grown plants of F2, BC3F1 and BC2F2 progenies using the CTAB (cetyl trimethyl ammonium bromide) method. In vitro DNA amplification through PCR was performed in a reaction volume of 25 μl, containing 50–75 ng template DNA, 100 μM of each dNTP, 1×PCR reaction buffer (10 mM Tris/HCl+50 mM KCl+0·01%, w/v, gelatin, pH 8·3), 1·5 mM MgCl2, 0·25 μM of each forward and reverse primers and 1·0 unit of Taq polymerase. The PCR amplification involved the following thermal profile: initial denaturation at 94°C for 4 min followed by 35 cycles of denaturation at 94°C for 1 min, annealing at 50–60°C (depending on the primer) for 1 min and extension at 72°C for 2 min followed by final extension at 72°C for 7 min. A negative control, without template DNA, was included in each plate during every amplification reaction. The amplification products were resolved on 2·5% high-resolution agarose (Amresco) prepared in 0·5×TBE buffer. After electrophoresis, gels were stained with ethidium bromide for 20 min, rinsed in deionized water and photographed under ultraviolet light using the gel documentation system (Fotodyne). Parental polymorphism and bulked segregant analysis (BSA) (Michelmore et al., Reference Michelmore, Paran and Kesseli1991) were taken up simultaneously. In this, equal quantity of DNA from ten highly resistant and ten highly susceptible plants from F2 population of the cross PR114/O. nivara acc. 81825 was bulked to generate resistant bulks (RB) and susceptible bulks (SB), respectively. In vitro DNA amplification of parents and the RB and SB was done using SSR markers selected at an interval of 5–10 centiMorgans (cM), from each of the 12 linkage groups presented by Temnykh et al. (Reference Temnykh, DeClerck, Lukashova, Lipovich, Cartinhour and McCouch2001). Once the chromosomal region harbouring the BB resistance gene was identified, based on BSA, the exact position of the gene was determined by analysing individual plants in segregating BC3F1 and BC2F2 progenies with polymorphic SSR markers selected from Temnykh et al. (Reference Temnykh, DeClerck, Lukashova, Lipovich, Cartinhour and McCouch2001) and IRGSP (2005 – Supplementary Table 18).
(iv) Data analysis and linkage mapping
Goodness of fit for all the loci to an expected 1:1 (in BC3F1) or 1:2:1 (in BC2F2) was tested using χ2 analysis. Linkage analysis of polymorphic markers was performed with the software package Mapmaker (Lander et al., Reference Lander, Green, Abrahamson, Barlow, Daly, Lincoln and Newburg1987) for f2 backcross and f2 intercross for BC3F1 and BC2F2 progenies, respectively. Recombination frequencies were converted to cM using Kosambi's mapping function (Kosambi, Reference Kosambi1944). Multipoint analysis was performed using an initial LOD (logarithm of the odds) threshold of 5·0 and maximum recombination fraction of 0·30. The polymorphic markers were ordered using the ‘Compare’ command. Additional markers were added afterwards on this frame using the ‘Try’ command. The final order was verified with the ‘Ripple’ command with a window size of 5 and LOD threshold of 10·0. The linkage map was finally drawn using the software MapChart, version 2.1 (Voorrips, Reference Voorrips2002), and graphical genotypes of individual plants were generated using the software GGT32 (Berloo, Reference Berloo1999; www.dpw.wageningen-ur.nl/pv/pub/ggt/).
(v) Quantitative trait locus (QTL) analysis
As presented above, the BB reaction was recorded as lesion length in cm, and plants in the segregating populations were classified as resistant or susceptible depending on whether lesion length was up to 5·0 cm or more. Although an alley was formed, both in BC3F1 and BC2F2 populations around the lesion length of 5·0 cm, still the variation was continuous. It was therefore analysed for QTL as well. The QTLs were detected and localized by single marker regression (SMA) and interval mapping (IM) using MapManager QTXb20 software (Manly et al., Reference Manly, Cudmore and Meer2001). In this analysis, the lesion length data of the individual plants were entered along with their genotypic data. The ‘marker regression’ function (P=0·01) was used to detect possible single marker loci associated with the QTL. Thereafter, the IM was used for localization and estimation of effects of the QTL. Significant (P<0·05) and highly significant (P<0·01) threshold levels were determined by the permutation test function of MapManager, which is based on the statistical methods developed by Churchill & Doerge (Reference Churchill and Doerge1994). A likelihood ratio statistic (LRS) value of 4·6 is equivalent to 1 LOD. Phenotypic variance (R 2) explained by the QTL was estimated as difference between the total variance and the residual variance expressed as the percentage of the total variance.
3. Results
(i) Inheritance of BB resistance in O. nivara acc. 81825
The O. nivara acc. 81825 showed resistance to all the seven Xoo pathotypes tested at maximum tillering stage. It was resistant to pathotype VII at seedling stage as well. Thus, the resistance gene in O. nivara acc. 81825 is effective at all the growth stages. Inheritance of BB resistance in O. nivara acc. 81825, based on an F2 progeny of the cross PR114/O. nivara acc. 81825, was reported earlier (Kaur et al., Reference Kaur, Grewal, Das, Vikal, Singh, Bharaj, Sidhu and Singh2005). Since our objective was simultaneous mapping and transfer of the gene to cultivated rice and preliminary evidence of segregation distortion for several SSR loci in the F2 progeny of the cross PR114/O. nivara acc. 81825, we generated BC2F2, BC2F3, BC3F1 and BC3F2 progenies for confirming the inheritance and mapping of the gene. The average lesion length in BC3F1 ranged from <0·5 to 20·4 cm. Of the 74 BC3F1 plants, 36 were resistant and 38 susceptible (Table 2), which fitted well to the expected 1:1 segregation (χC2=0·014, Table 2). In BC2F2, the average lesion length ranged from 0·5 to 24·2 cm (Fig. 1). Of the 396 BC2F2 plants, 290 showed resistant reaction, whereas 106 showed susceptible reaction (Table 2), which again fitted well to one-gene segregation (χC2=0·569). Of the 396 BC2F3 progenies, 115 were homozygous resistant, 175 were segregating and 106 were homozygous susceptible, which fitted well to the expected 1:2:1 segregation (Table 2). Similarly, out of 1095 BC3F2 plants, 809 were resistant and 289 susceptible, which again fitted well to 3:1 (Table 2) expected of a single gene segregation. Thus, the BB resistance gene in O. nivara acc. 81825 behaves as a single dominant gene even when transferred to cultivated rice O. sativa.
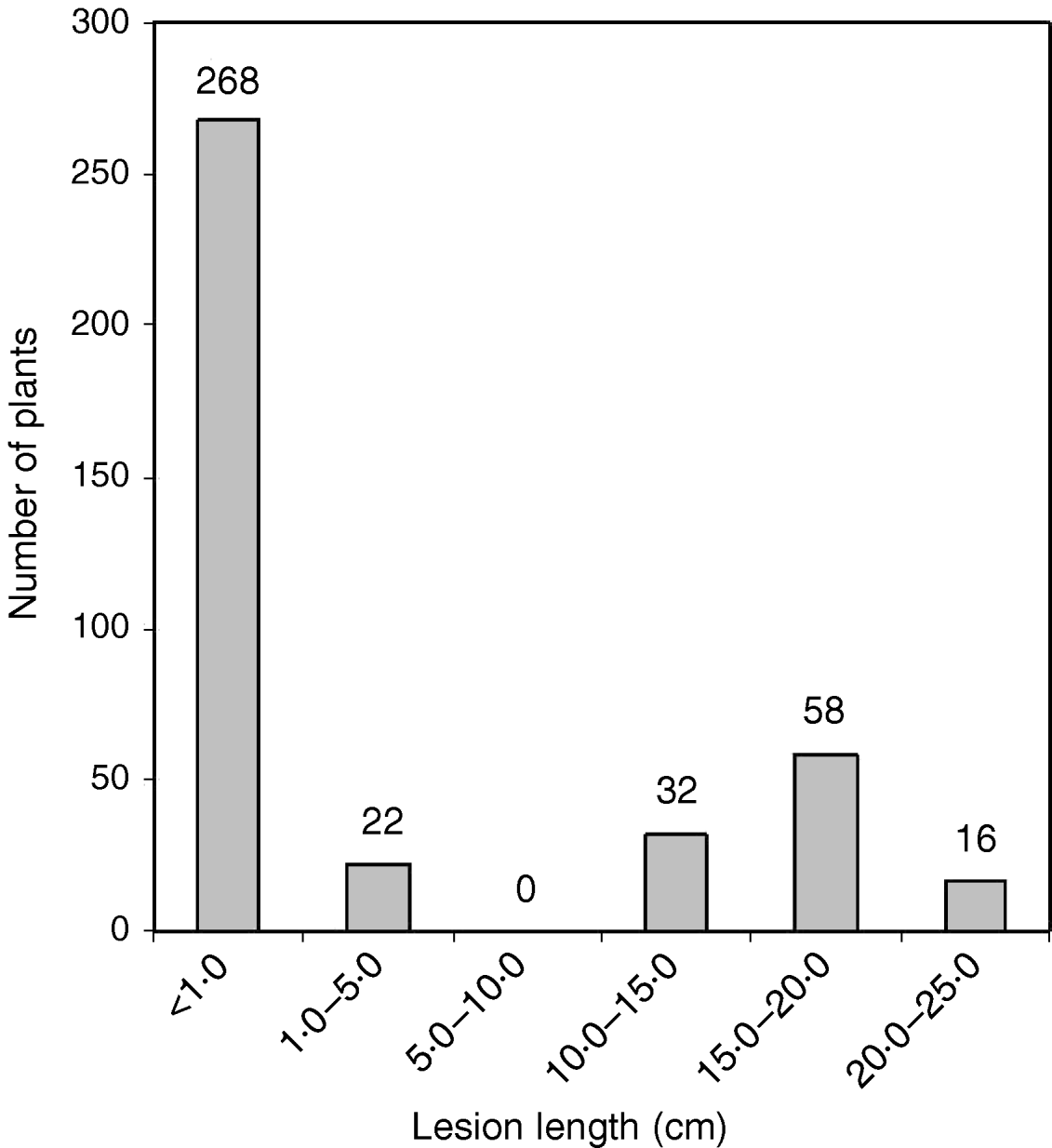
Fig. 1. The distribution of average lesion length in BC2F2 progeny of the cross PR114/O. nivara acc. 81825/2*PR114. The average lesion lengths of O. nivara acc. 81825 and PR114 were 0·5 and 16·4 cm, respectively.
Table 2. Frequency of resistant and susceptible plants in the BC3F1, BC3F2, BC2F2 and BC2F3 populations
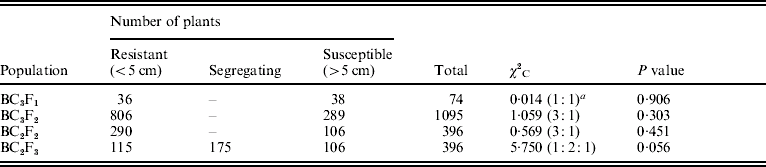
a Ratios in parentheses are the ones expected, based on one-gene segregation. The average lesion length in PR114 and O. nivara acc. 81825 against pathotype VII was 16·4 and 0·5 cm, respectively.
(ii) Parental polymorphism and BSA
Parental polymorphism and BSA (Michelmore et al., Reference Michelmore, Paran and Kesseli1991) were conducted simultaneously. SSR markers from the map of Temnykh et al. (Reference Temnykh, DeClerck, Lukashova, Lipovich, Cartinhour and McCouch2001) were selected at an interval of 5–10 cM, from all the 12 linkage groups. A total of 402 SSR markers were tested for parental polymorphism and BSA. Out of the total 402 SSR markers tested, 211 (52·5%) were monomorphic and 191 (47·5%) were polymorphic when resolved in 3·0% high-resolution agarose gels. Parental polymorphism ranged from as low as 21·7% for chromosome 12 to 61·3% for chromosome 7. All the 402 markers were simultaneously tested for BSA using the DNA of RB and SB prepared from the F2 population of the cross PR114/O. nivara acc. 81825. Any marker that assorts independent of resistance gene will show both parent bands in RB as well as SB, whereas a marker linked to a dominant resistance gene in coupling phase will show only susceptible parent band in the SB, but in RB both resistant and susceptible parent bands will appear, though with varied intensity. Of the 191 polymorphic markers, as many as 48 markers showed distorted segregation based on BSA, i.e. both the RB and SB had either PR114 allele or O. nivara allele only (Fig. 2). Thirteen of the distorted markers map on chromosome 5. Markers RM567, RM127, RM317 and RM252 located on distal end of chromosome 4L showed the banding pattern expected of a marker linked to the target gene (Fig. 3). RM317 showed only susceptible parent band in SB and resistant parent band in RB, although RB was expected to have bands from both the parents. Markers RM252, RM537 and RM551 that map distal to RM317, towards the centromere (Temnykh et al., Reference Temnykh, DeClerck, Lukashova, Lipovich, Cartinhour and McCouch2001), also showed distorted segregation. The region bracketed by RM317 and RM567 was about 35 cM (Fig. 4a, Temnykh et al., Reference Temnykh, DeClerck, Lukashova, Lipovich, Cartinhour and McCouch2001).
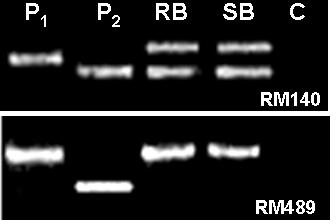
Fig. 2. Banding pattern of parents, RB and SB for RM140 and RM489. RM140 shows both parent bands in RB as well SB, whereas RM489 shows only P1 (O. nivara) band, both in RB and SB. P2 is O. sativa cv PR114 and C is control (no DNA).

Fig. 3. Bulk segregant analysis showing association of RM567, RM127, RM317 and RM352 with BB resistance gene in O. nivara. P1 is O. nivara, P2 is PR114 and C is control (no DNA). SB shows the presence of PR114 band only, whereas the RB shows the presence of bands from both the parents as expected of a dominant gene, except for RM317 wherein RB had only O. nivara band.

Fig. 4. Linkage map of chromosome 4L showing map position of Xa30(t). (a) Linkage map of 4L based on Temnykh et al. (Reference Temnykh, DeClerck, Lukashova, Lipovich, Cartinhour and McCouch2001) ; (b) linkage map of 4L based on 74 BC3F1 plants and (c) based on 336 BC2F2 plants. BSA identified markers RM317, RM127 and RM567 to be associated with the BB resistance gene.
(iii) Mapping of BB resistance gene in O. nivara acc. 81825
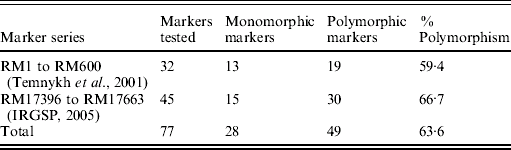
Although the BSA was based on F2 progeny of the cross PR114/O. nivara acc. 81825, mapping was done using BC3F1 and BC2F2 progenies due to distorted segregation for several SSR loci as described above. For chromosome 4, 32 SSR markers were tested for polymorphism from the map of Temnykh et al. (Reference Temnykh, DeClerck, Lukashova, Lipovich, Cartinhour and McCouch2001) and 45 from IRGSP (2005) (Table 3). Out of the total 77 SSR markers tested, 49 (63·6%) were polymorphic (Table 3). Of 49 polymorphic markers, 34 were analysed on 74 BC3F1 plants. Twenty-one of these showed expected 1:1 segregation for homozygotes (for PR114 allele) and heterozygotes and 13 showed significant deviation from the expected 1:1, with χ2 values ranging from 4·4 to 46·4. All the markers that showed segregation distortion had PR114 allele in higher frequency and map towards the centromere, whereas the ones that did not show the distortion map towards telomere.
Table 3. Parental polymorphism for SSR markers between PR114 and O. nivara acc. 81825 in chromosome 4
Analysis of 74 BC3F1 plants for BB reaction and genotypic data for 34 SSR markers, using the software package Mapmaker, mapped BB resistance gene from O. nivara between RM17499 and RM17502, which are 4·0 cM apart (Fig. 4b). Only 26 of the 34 markers could be mapped using this population. Markers RM252, RM537 and RM551 that showed segregation distortion in favour of PR114 allele did not show linkage with other markers but formed a separate linkage group (data not shown). The linkage map thus generated with 26 SSR markers spans 27·3 cM region (Fig. 4b), which is almost similar to map distance between RM317 and RM567 (Fig. 4a) as reported by Temnykh et al. (Reference Temnykh, DeClerck, Lukashova, Lipovich, Cartinhour and McCouch2001). Since the BB resistance gene showed co-segregation with RM17499, an additional 336 BC2F2 plants were used for fine mapping of the gene. Six SSR markers, RM17480, RM17483, RM17488, RM17496, RM17499 and RM17502 (IRGSP, 2005; Supplementary Table 18), were analysed in 336 BC2F2 plants. All these markers showed expected 1:2:1 frequency of individuals homozygous for PR114 alleles, heterozygous, and homozygous for O. nivara allele, respectively. Mapmaker analysis showed the BB resistance gene to map between RM17499 and RM17502, which are about 176 kb apart and flank the BAC clone OSJNBb0085C12. To further narrow down the region, 14 SSRs were identified from the BAC clone OSJNBb0085C12 and their primers designed. However, none of the 14 SSRs showed polymorphism between the parental lines. Subsequently, 17 gene-based primers were designed from 12 loci, including five nucleotide-binding site–leucine-rich repeat (NBS–LRR) loci, annotated from BAC clone OSJNBb0085C12 (http://rice.plantbiology.msu.edu/cgi-bin/gbrowse/rice/). Primers designed from two loci, LOC_Os04g53060 (5′-TGGAAACAAGGAAGGTTTCG-3′F and 5′-TGGACTGAGATGAGGTGCTG-3′R) and LOC_Os04g53120 (5′-ATGAATGGATTCGCCAGAT-3′F and 5′-CGTCTCTTCAGCCTCAAACC-3′R), were polymorphic, whereas the other 15 markers were monomorphic even after restriction with several restriction enzymes for which restriction sites were present in the Nipponbare genome. The locus LOC_Os04g53060 showed amplification in cultivated parent line PR114 but no amplification in O. nivara, whereas the locus LOC_Os04g53120 showed amplification in both the parents. Analysis of 336 BC2F2 plants with six SSR and two gene-based markers showed that the gene maps between LOC_Os04g53060 and LOC_Os04g53120 at a distance of 0·2 cM from LOC_Os04g53060 and 0·9 cM from LOC_Os04g53120 (Fig. 4c). Thus the BB resistance gene from O. nivara maps on chromosome 4L in 1·1 cM region bracketed by annotated loci LOC_Os04g53060 and LOC_Os04g53120 (http://rice.plantbiology.msu.edu/cgi-bin/gbrowse/rice/).
(iv) Mapping BB resistance as QTL
The BB resistance gene could be mapped as a major gene after classifying the plants as resistant or susceptible, based on lesion length, as presented in the preceding section. The BC3F1 population was also analysed for QTL, based on lesion length. Both SMA and IM were used for identifying the regions associated with BB resistance. Markers that showed co-segregation were removed as redundant loci by the software for analysis. With IM, the region between markers RM17499 and RM17502 had the highest LRS value of 194·6 (which equates to a LOD value of 42·0) and it explained 93% of the total phenotypic variation (Fig. 5). The threshold LRS value (P<0·001), based on 1000 permutations, was 18·1. Thus the QTL for BB resistance mapped on chromosome 4L is highly significant. Similarly, IM of the BC2F2 population of 336 plants identified an interval between markers LOC_Os04g53060 and LOC_Os04g53120 as having the highest LRS value, being 460 (which equates to a LOD score of 100·0), and it again explained 93% of the total phenotypic variation.
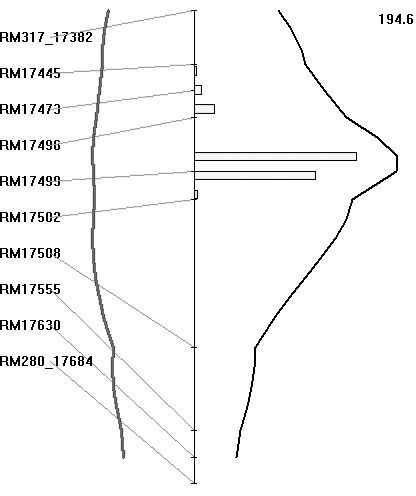
Fig. 5. Map location of BB resistance QTL transferred from O. nivara. A partial linkage map of chromosome 4L is shown along. Curves in black represent the LRS values for the markers and curves in grey represent the regression coefficients (additive effects). The numerical value on top of the figure is the highest LRS values for the QTLs detected on chromosome 4L. Histogram represents estimates of confidence interval by bootstrap resampling.
(v) Graphical genotypes of the BC3F1 and BC2F2 plants
Graphical genotypes were generated for all the 74 BC3F1 plants for 27·3 cM region using 26 SSR markers and for 336 BC2F2 plants using six SSR markers. Among the 36 resistant BC3F1 plants, the smallest introgression, around the BB resistance gene, was observed in plant no. 1358-15 for the chromosomal region spanning SSR markers RM17473 and RM17499 (Fig. 6a), which is ∼476 kb. Similarly out of 336 BC2F2 plants, graphical genotype of four resistant and three susceptible plants are presented in Figure 6b. All the resistant plants had introgression in the region spanning the loci LOC_Os04g53060 and LOC_Os04g53120. Plant no. 1360-105 was resistant but did not show any introgression for RM17502 thus ruling out the possibility of this marker associated with BB resistance. Plant no. 1360-54 was resistant and had introgression for LOC_Os04g53060, LOC_Os04g53120 and RM17502 but did not show introgression for RM17499, thereby indicating that RM17499 is also not associated with BB resistance. Graphical genotypes of seven BC2F2 plants presented in Figure 6b thus confirm the mapping data, which showed BB resistance gene from O. nivara being located between LOC_Os04g53060 and LOC_Os04g53120, which is about 38·4 kb.

Fig. 6. Graphical genotype for chromosome 4L of a BB-resistant BC3F1 plant (no. 1358-15) (a) and seven BC2F2 plants (b). Numbers below bars in (b) are the BC2F2 plant numbers; 1360-7, 1360-105, 1360-20 and 1360-54 were resistant to BB, whereas 1360-134, 1360-46 and 1360-5 were susceptible. Black region indicates introgression from O. nivara chromatin homozygous condition; dark grey as heterozygous regions and light grey indicates PR114 chromatin. Numerical values on the right in (a) and the values adjoining the markers in (b) are the map distances in cM.
(vi) Transfer of BB resistance gene from O. nivara into O. sativa cv PR114
Out of 74 BC3F1 plants, 36 were resistant and 38 susceptible. Lesion length in resistant plants ranged from <1·0 to 3 cm with 31 plants having less than 1 cm lesion length. Since graphical genotypes were generated for all the BC3F1 plants, plant no. 1358-15 was identified as having the smallest introgression around the region harbouring BB resistance. This plant was self-pollinated and, based on flanking markers linked to the BB resistance gene, homozygous BC3F2 plants were identified from its progeny. These plants were resistant with lesion length less than 1 cm (Fig. 7). The BC3F3 progenies that were homozygous for BB resistance gene and phenotypically like PR114 have been identified and are available for use in breeding programmes.

Fig. 7. BB reaction of PR114 (2), O. nivara acc. 81825 (3) and a BC3F2 plant (4) when inoculated at maximum tillering stage with Xoo pathotype VII. Sample 1 is uninoculated leaf of PR114.
Discussions
(i) Polymorphism and segregation distortion
Overall, 47·5% polymorphism was observed between O. sativa cv PR114 and O. nivara acc. 81825 for all the 12 chromosomes. McCouch et al. (Reference McCouch, Teytelman, Xu, Lobos, Clare, Walton, Fu, Maghirang, Li, Xing, Zhang, Kono, Yano, Fjellstrom, DeClerck, Schneider, Cartinhour, Ware and Stein2002) observed 86% polymorphism between a japonica cultivar, Nipponbare, and an indica cultivar, Kasalath, when they resolved a set of 545 SSR markers in 3·0% agarose gel, whereas He et al. (Reference He, Li, Zhu, Tan, Zhang and Lin2006) observed only 17% polymorphism between two indica lines, Zhenzhuai and IR24. The polymorphism level observed between the parental lines PR114 and O. nivara acc. 81825, although not as high as in the indica–japonica crosses, seems sufficiently high for map-based cloning of the BB resistance gene from O. nivara. Out of 191 polymorphic SSR markers tested in parents and the RB and SB, 48 markers showed segregation distortion wherein both the RB and SB had only either PR114 allele or O. nivara allele. The phenomenon of segregation distortion has been frequently reported in wide crosses in many crop species and is well documented in intraspecific and interspecific crosses in rice (Xu et al., Reference Xu, Zhu, Xiao, Huang and McCouch1997; Lorieux et al., Reference Lorieux, Ndjiondjop and Ghesqui2000; Heuer & Miézan, Reference Heuer and Miézan2003). The preferential transmission or segregation distortion might be due to gamete elimination, pollen killer genes and the phenomena of restriction in recombination as well. The work of Nakagahra et al. (Reference Nakagahra, Omura and Iwata1972) led to the suggestion that the gametophytic loci were responsible for the partial or total elimination of gametes carrying one of the parental alleles. Although 48 loci showed segregation distortion in this cross, the region harbouring the BB resistance gene in O. nivara showed normal segregation. However, markers RM252, RM551 and RM537 that map on 4L towards the centromere (Temnykh et al., Reference Temnykh, DeClerck, Lukashova, Lipovich, Cartinhour and McCouch2001) showed distortion in favour of O. sativa allele. Sterility gene S28(t) from Oryza glumaepatula has been mapped on chromosome 4 (Sobrizal et al., Reference Sobrizal and Yoshimura2002) in the same region where the above mentioned SSR markers map. In crosses between O. sativa and O. glumaepatula, the sterility gene S28(t) causes elimination of O. sativa allele (Sobrizal et al., Reference Sobrizal and Yoshimura2002), but in the present cross, O. sativa alleles were favoured.
(ii) BB resistance gene in O. nivara acc. 81825 is novel
The BB resistance gene under study was mapped to ∼176 kb region on chromosome 4L using BC3F1 population. Four other BB resistance genes, Xa1, Xa2, Xa12 and Xa14, are also reported on 4L (Ogawa et al., Reference Ogawa, Morinaka, Fujii and Kimura1978; Taura et al., Reference Taura, Ogawa, Tabien, Khush, Yoshimura and Omura1987; Yoshimura et al., Reference Yoshimura, Yamanouchi, Katayose, Tok, Wang, Kono, Kurata, Yano, Iwata and Sasaki1998; He et al., Reference He, Li, Zhu, Tan, Zhang and Lin2006), but none of these are effective against any of the seven pathotypes prevalent in Punjab (Table 1), and hence allelic test is not possible. The Xa1 was closely linked to Xa12 and Xa2 with Xa1 mapping towards the centromere and Xa2 towards the telomere with a recombination distance of 4·0 cM between them (Kinoshita & Takashi, Reference Kinoshita and Takashi1991). Physical mapping and gene sequencing, however, placed Xa2 towards the centromere and Xa1 towards telomeric region (Yoshimura et al., Reference Yoshimura, Yamanouchi, Katayose, Tok, Wang, Kono, Kurata, Yano, Iwata and Sasaki1998; He et al., Reference He, Li, Zhu, Tan, Zhang and Lin2006). The Xa2 has been mapped to a 190 kb region on overlapping BAC clones OSJNBa0008M17, OSJNBa0093O08 and OSJNBa0058K23 (He et al., Reference He, Li, Zhu, Tan, Zhang and Lin2006). The Xa1 gene (Os04g0622600) annotated in RAP-DB (http://rapdb.dna.affrc.go.jp/cgi-bin/gbrowse/IRGSP40/) corresponds to gene LOC-Os04g53160, whose co-ordinates span the genome sequences 31 437 628 bp to 31 443 630 bp (http://rice.plantbiology.msu.edu/cgi-bin/gbrowse/rice/). Both major gene and QTL mapping data and the graphical genotypes of BC3F1 and BC2F2 plants show that the gene described in the present study maps between markers LOC_Os04g53060 (start position 31 386 340 bp) and LOC_Os04g53120 (end position 31 425 732 bp), which are about 38·4 kb apart and cover the BAC clone OSJNBb0085C12. The BAC clone OSJNBb0085C12 harbours five NBS-LRR disease resistance gene motifs, viz. LOC_Os04g52970 (31334008–31339348), LOC_Os04g53000 (31346084–31347211), LOC_Os04g53050 (31381060–31385140), LOC_Os04g53060 (31386340–31388283) and LOC_Os04g53160 (31437628–31443630). In addition, LOC_Os04g53120 (31419036–31425732), a putative NB-ARC-domain-containing protein, is also present on this BAC clone. The Xa1 locus corresponds to LOC_Os04g53160 (31437628–31443630), whereas Xa2 may correspond to the locus LOC_Os04g52780 (31201695–31206034), which maps on another BAC clone OSJNBa0058K23 and is an LRR-receptor protein kinase as postulated by He et al. (Reference He, Li, Zhu, Tan, Zhang and Lin2006). None of the four BB resistance genes, Xa1, Xa2, Xa12 and Xa14, on 4L are effective against any of the seven pathotypes prevalent in Punjab, but the gene identified in the present study is resistant against all the seven pathotypes. All the evidence suggests that the gene is novel. High-resolution mapping around this locus is in progress, and final characterization will be possible after cloning and sequencing of the gene. This gene, therefore, is tentatively designated Xa30(t). The homozygous BB-resistant BC3F3 progenies in the background of PR114 will serve as a source of this gene. The closely linked SSR marker RM17499 and LOC_Os04g53120 could be used as diagnostic markers for marker-assisted breeding and gene pyramiding as the locus LOC_Os04g53160 is not amplified in O. nivara. Since no known dominant BB resistance gene is effective against prevalent pathotypes in North Western India, the new gene could be useful for development of BB-resistant F1 hybrids and varieties. Locus LOC_Os04g53120 is a putative NB-ARC-domain-containing protein and is a part of the locus Xa1 along LOC_Os04g53160 (http://rapdb.dna.affrc.go.jp/cgi-bin/gbrowse/IRGSP40/) that has functions similar to Xa1. Therefore, locus LOC_Os04g53060 (=Os04g621900 – RAP-DB), which is an NBS-LRR and is a 1944 bp long gene, is a candidate gene for Xa30(t).
Plant disease resistance genes frequently occur in the form of a tandem repeat multigene family. Four BB resistance genes, Xa3, Xa26(t), Xa4 and Xa22, have been mapped to the terminal region of the long arm of chromosome 11(Lin et al., Reference Lin, Zhang, Xie, Gao and Zhang1996; Yoshimura et al., Reference Yoshimura, Umehara, Kurata, Nagamura, Sasaki, Minobe and Iwata1996; Sun et al., Reference Sun, Yang, Wang and Zhang2003; Yang et al., Reference Yang, Sun, Wang and Zhang2003). All these genes are tightly linked and may belong to a multigene family (Yang et al., Reference Yang, Sun, Wang and Zhang2003). BB resistance genes Xa1, Xa2, Xa12, Xa14 and Xa30(t), which map in ∼600 kb region on chromosome 4L, may also belong to a multigene family. The chromosomal region between RM17499 and RM17502 where Xa30(t) maps is around 176 kb and shows a recombination rate of ∼60 kb/cM, which is high to moderately high as reported in certain other high recombination regions in rice (Wu et al., Reference Wu, Mizuno, Hayashi-Tsugane, Ito, Chiden, Fujisawa, Katagiri, Saji, Yoshiki, Karasawa, Yoshihara, Hayashi, Kobayashi, Ito, Hamada, Okamoto, Ikeno, Ichikawa, Katayose, Yano, Matsumoto and Sasaki2003). The high frequency of recombination in resistance-gene-rich regions may be the cause for the generation of new resistance gene specificities.
This project was carried initially with financial support from the Rockefeller Foundation and later with financial support from the Department of Biotechnology, Ministry of Science and Technology, Government of India (project no. BT/AB/03/FG-2/2003). The O. nivara acc. 81825 was procured from IRRI, Philippines, and we gratefully acknowledge their support. Our thanks to Dr Parveen Chhuneja for her help in QTL analysis.












