Introduction
Fetal growth restriction (FGR) is a serious complication of pregnancy, often followed by altered brain structure, cognitive deficits, motor disability, and neuropsychological disorders. Reference Miller, Huppi and Mallard1 Several genetic and environmental factors may induce FGR, the most common being placental dysfunction, which leads to abnormal maternal–fetal exchange and fetal hypoxia. Reference Nardozza, Caetano and Zamarian2 Compensatory fetal hemodynamic redistribution with cerebral vasodilation attempts to spare the fetal brain from hypoxic damage, which is known as fetal brain-sparing. Reference Giussani3 It is still under debate whether and how this may benefit the developing brain.
In the same cohort of FGR children as used in this study, we have previously observed that the sonographic presence of fetal brain-sparing was associated with higher neonatal cerebral oxygen saturations. Reference Tanis, Boelen and Schmitz4 At follow-up, fetal brain-sparing and higher cerebral oxygen saturations were related to better behavior and executive functioning at 4 years of age. Reference Richter, Salavati and Kooi5 However, high cerebral oxygen saturation levels have also been associated with poorer cognition, in particular, a poorer Performance Intelligence Quotient (IQ). Reference Richter, Salavati and Kooi5,Reference Verhagen, Van Braeckel and van der Veere6
There is evidence that the early-life environment influences long-term neurodevelopment through epigenetic mechanisms. Reference Stroud, Su and Hrvatin7,Reference Schachtschneider, Welge and Auvil8 Among these mechanisms, methylation of cytosine-phosphate-guanine (CpG) dinucleotides within promoter regions contributes to maintaining long-lasting states of gene repression. Reference Weber, Hellmann and Stadler9 Oxygen levels have shown to alter DNA methylation and directly affect the activity of transcription factor hypoxia-inducible factor-1α (HIF1α), which has popular target genes exerting neurotrophic functions. Reference Bernaudin, Nedelec, Divoux, MacKenzie, Petit and Schumann-Bard10–Reference Mutoh, Sanosaka, Ito and Nakashima13
The aim of this study was therefore to analyze whether in our FGR cohort, fetal brain-sparing was associated with altered methylation at predefined loci of the genes encoding HIF1α, its neurodevelopmentally important target genes (vascular endothelial growth factor A [VEGFA], erythropoietin [EPO], brain-derived neurotrophic factor [BDNF], and neurotrophic tyrosine kinase, receptor, type 2 [NTRK2]) and the EPO-receptor gene (EPOR) at 4 years of age. We used buccal DNA as it is convenient to sample at this age and has been demonstrated to show methylation patterns comparable with neuronal DNA. Reference Smith, Kilaru and Klengel14 We further explored whether differential methylation of these genetic loci was related to neurodevelopmental outcome (intelligence, behavior, and executive functions) at this age, which may explain our previous findings.
Method
Study design and population
This was a prospective observational cohort study including 21 children born following FGR between June 2012 and May 2014 at the University Medical Center Groningen (UMCG), The Netherlands.
Antenatal inclusion was based on FGR diagnosed by a fetal abdominal circumference or estimated fetal weight below the 10th percentile or by flattening of the fetal growth curve by more than 30 percentiles compared with the preceding examination. Exclusion criteria were structural or chromosomal abnormalities, multiple pregnancy, or evidence of intrauterine infection. At the age of 4 years, surviving children with complete perinatal hemodynamic assessment (antenatal cerebroplacental Doppler and postnatal cerebral oxygenation measurements), consent for follow-up, and sufficient knowledge of the Dutch language received neurodevelopmental testing. Buccal DNA was sampled during this visit. Participants declining DNA sampling were excluded for the purpose of this study.
We chose to assess the children at the age of 4 years, as children enter school at this age in The Netherlands and are able to participate in the Wechsler Preschool and Primary Scale of Intelligence (WPPSI) IQ test. Written informed parental consent for participation was obtained and the study was approved by the Medical Ethical Committee of the UMCG.
Perinatal data
We prospectively collected gestational data (parental ethnicity, maternal age, body mass index [BMI], smoking habits, antidepressants, pregnancy complications, antenatal steroids, and magnesium sulfate), fetoplacental data (Doppler measurements and placental histology as examined by a perinatal pathologist according to criteria applicable at the time of examination Reference Lewis and Perrin15–Reference Bendon26 ), and neonatal data (gestational age, delivery mode, sex, birth weight, head circumference, Apgar score, admission to the neonatal intensive care unit, postnatal complications, the need for steroids, and cerebral oxygen saturation as measured with near-infrared spectroscopy during first three days after birth).
Fetal brain-sparing
Antenatal Doppler sonography was performed to assess the pulsatility index (PI) of the umbilical and middle cerebral artery. The cerebroplacental ratio (CPR) was calculated by dividing the latter by the first. Reference Gramellini, Folli, Raboni, Vadora and Merialdi27 A CPR below 1 was considered as evidence for fetal brain-sparing. The measurements were performed at least once a week upon diagnosis of FGR. The last measurement before birth was included for analysis.
Methylation of neurodevelopmental genes at 4 years of age
Gene selection
Based on our hypothesis, we chose to analyze oxygen-dependent regulatory genomic regions encoding HIF1α and well-known neurotrophic factors, including EPO, VEGFA, and BDNF, which play important roles during brain development and contain binding sites for HIF1α. Reference Buemi, Cavallaro and Floccari28–Reference Gottmann, Mittmann and Lessmann30 The selected DNA sequences with its CpG positions and relevant transcription factor binding sites are presented in Fig. 1. We selected the promoter region of HIF1A, which encodes HIF1α. This region contains a hypoxia-response element (HRE), to which HIF1α is able to bind and increase expression of HIF1α under hypoxia in an autoregulatory fashion. Reference Koslowski, Luxemburger, Türeci and Sahin31 Next to the HRE lies a binding site for the transcription factor Kaiso (also known as ZBTB33), which has been suggested to repress HIF1α expression. Reference Pierre, Longo and Bassey-Archibong32 We further selected the promoter and the distal enhancer region of EPO, which contain binding sites for HIF1β and HIF1α, respectively, and together are responsible for the expression of EPO under hypoxic conditions. Reference Steinmann, Richter and Dammann33,Reference Dewi and Fatchiyah34 In addition, the promoter region of EPOR was analyzed. EPOR does not contain any known HREs, but the selected region has been implicated in developmental downregulation of EPOR expression in the brain. Reference Wallach, Zhang and Hartmann35 We further selected an HRE locus in the promoter region of the gene encoding VEGFA. Reference Sundrani, Reddy and Joshi36,Reference Pisani, Cammalleri and Dal Monte37 Likewise, an HRE locus within the promoter region of NTRK2 was selected, which encodes Tropomyosin receptor kinase B (TrkB), a receptor for BDNF and other neurotrophins. Reference Martens, Kirschner, Warnecke and Scholz11 In addition, we selected a region within the promoter of BDNF exon 4. This region contains two binding sites for the transcription factor cyclic adenosine monophophate response element binding protein (CREB), which has shown to mediate neuroprotective effects of EPO in cerebral ischemia. Reference Fang, Chartier and Sodja38–Reference Kundakovic, Gudsnuk, Herbstman, Tang, Perera and Champagne41

Fig. 1. DNA sequence to analyze before bisulfite treatment depicting CpG positions (bolded and numbered in the direction of sequencing) and important transcription factor binding sites (underlined with transcription factors in grey boxes). BDNF, brain-derived neurotrophic factor; CpG, 5′-cytosine-phosphate-guanine-3′ dinucleotide; CREB, cyclic adenosine monophosphate response element binding protein; EPO, erythropoietin; EPOR, erythropoietin receptor; Ets, E26 transformation-specific; HIF1A/HIF1α, hypoxia-inducible factor-1 alpha; HIF1β, hypoxia-inducible factor-1 beta; NTRK2, neurotrophic tyrosine kinase, receptor, type 2; Sp1, specificity protein 1; VEGFA, vascular endothelial growth factor A.
DNA sampling and isolation
Buccal cells were collected during follow-up using Isohelix Buccal Swabs (Cell Projects Ltd, Kent, UK). All samples were stored at 4°C until DNA isolation. Once all samples were collected, isolation of DNA was performed according to the protocol of the BuccalFix Plus DNA Isolation Kit (Cell projects Ltd, Kent, UK). Quality and concentration of the isolated DNA were checked by gel electrophoresis and the NanoDrop 2000c spectrophotometer (Thermo Fisher Scientific, Waltham, MA). Isolated DNA was subsequently stored at −80°C until bisulfite treatment.
Analysis of DNA methylation by pyrosequencing
Before pyrosequencing, 250 ng of isolated DNA was treated with bisulfite to convert unmethylated cytosine residues into uracil leaving only methylated cytosine residues. This was done using the EZ DNA Methylation-Gold Kit (Zymo Research, Irvine, CA) according to the manufacturer’s protocol with a discard of the flow-through and an extra round of 30 s centrifuging at full speed after step 8. For polymerase chain reaction (PCR), we prepared a mastermix containing 12.5 µl HotStarTaq DNA Polymerase, 10.5 µl of sterile water, and a 1 µl of forward and reverse primer mix (each 10 µM) per 1 µl bisulfite template. A negative control without template was included to check for contamination. We used the T100 Thermal Cycler (Bio-Rad, Hercules, CA) and the following conditions for PCR amplification: 95°C for 15 min, 45 cycles of 94°C for 30 s, assay specific temperature for 30 s, 72°C for 30 s, followed by a final step of 72°C for 7 min. Assay specific temperatures: 56°C for HIF1A promoter and NTRK2; 58°C for EPO promoter, EPOR, and VEGFA; 62°C for EPO enhancer and BDNF exon 4. The modified DNA was then stored at −20°C.
PCR and pyrosequencing primers (Table 1) were designed using the PyroMark Assay Design software (Qiagen, Hilden, Germany). Primers used for HIF1A, VEGFA, and EPO were previously designed by Bekkering et al. Reference Bekkering, Leeuwerke and Tanis42 After PCR amplification of the DNA region of interest, pyrosequencing was performed using PyroMark Q24 and PyroMark Q48 Autoprep (Qiagen). Methylation levels of each CpG position (given as percentages) and the bisulfite conversion rate were analyzed using the PyroMark Q24 Software and PyroMark Q48 Advanced Software (Qiagen). The maximum allowed percentage of non-conversion was set at 5% in the analysis software. The actual non-conversion rate in the samples was between 0.5% and 3%, wherefore we considered the bisulfite conversion successful. Samples which failed quality control were replicated up to three times until quality control was passed. The sample was excluded if low quality persisted (Supplementary Table S1).
Table 1. PCR and sequencing primers, sequence to analyze after bisulfite treatment, and the respective genomic region per gene as based on the Homo Sapiens GRCh38.p 13 primary assembly
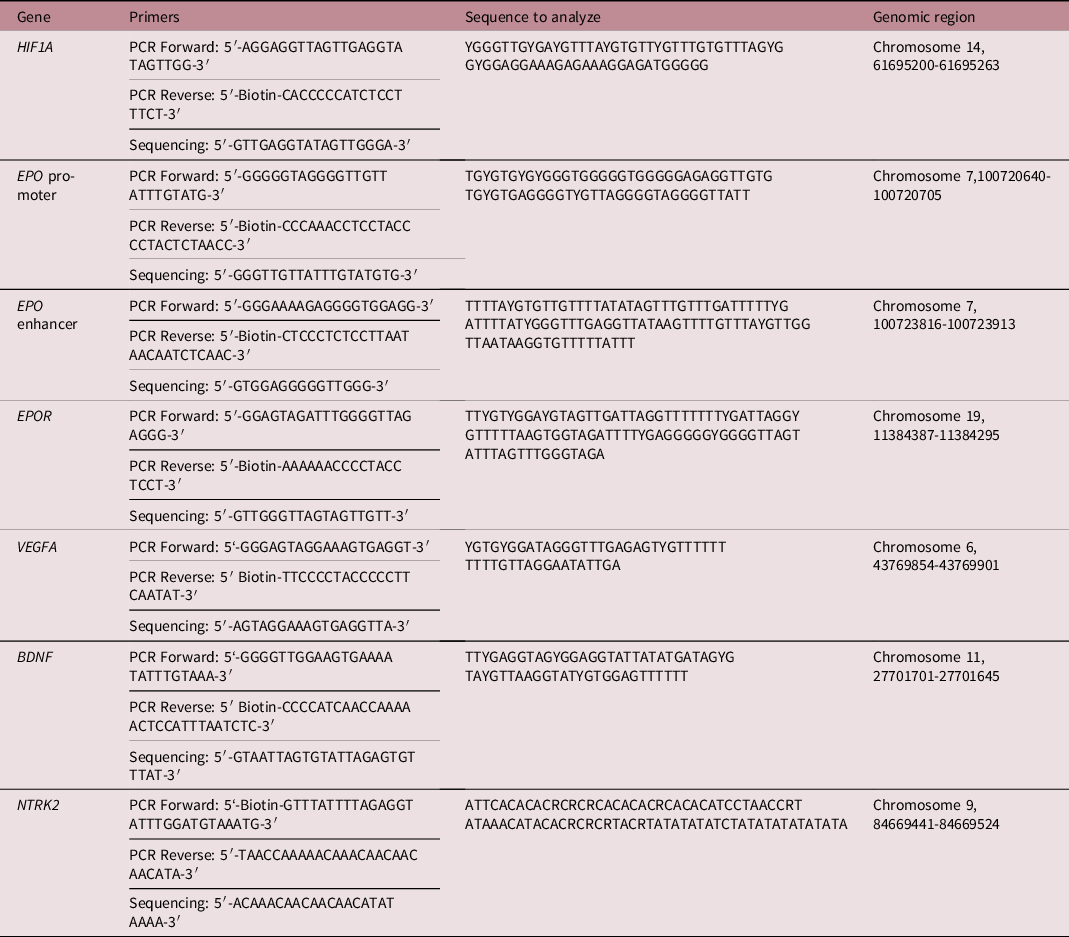
BDNF, brain-derived neurotrophic factor; EPO, erythropoietin; EPOR, erythropoietin receptor; HIF1A, hypoxia-inducible factor-1 alpha; NTRK2, neurotrophic tyrosine kinase, receptor, type 2; PCR, polymerase chain reaction; VEGFA, vascular endothelial growth factor A.
Neurodevelopmental outcome
Neurodevelopmental outcome was assessed at the age of 4 years based on cognition, behavior, and executive functions. Cognition was tested using the WPPSI for children aged 4–7 years (3rd edition, Dutch version), yielding a normed Full Scale, Verbal, and Performance IQ. Total, internalizing, and externalizing behavior (normed T-scores) were assessed using the Child Behavior Checklist for ages 1.5–5 years. Executive functions were examined using the Behavior Rating Inventory of Executive Function-Preschool Version for children aged 2–5 years. More specifically, inhibitory self-control, (emotional) flexibility, and emergent metacognition (problem solving using working memory and planning) were tested, which produced normed T-scores for the respective indices and total executive functioning.
Statistical analysis
The statistical software package SPSS 23.0 (IBM Corporation, Armonk, NY, USA) was used for analyses. A (two-sided) p-value below 0.05 was considered significant and a (two-sided) p-value between 0.05 and 0.1 a possible trend or tendency toward a significant association. As this was an explorative, hypothesis-forming study, we refrained from correcting our p-values to reduce the chance of type 2 error.
First, each variable was tested for normal distribution using the Shapiro–Wilk test. Second, differences in population characteristics and the percentage of methylation per CpG location between FGR children with and without evidence of fetal brain-sparing were tested using a Student’s t-test or Mann–Whitney U test, depending on normality of the data. To test for differences between two binary variables a chi-square test was used. In addition, to better understand how methylation of individual CpG sites within one region relate to each other and potential transcription factor binding sites, we performed correlation analyses between CpGs using Pearson or Spearman’s rank correlation analysis, depending on normality of the data. Third, cohort characteristics, which are likely to have a separate effect on DNA methylation, such as perinatal steroid use, gestational age, sex, child age and BMI (z-score) at DNA sampling, maternal smoking, medication, age, BMI, and parental ethnicity, were regarded as potential confounders. Reference Hogg, Price, Hanna and Robinson43–Reference Virani, Dolinoy and Halubai45 If in our population, these variables were significantly associated with both brain-sparing and CpG methylation, their confounding effect was adjusted for in a multivariate linear regression model. Finally, to secondarily assess whether differential CpG methylation was associated with neurodevelopmental outcome, correlation analyses were applied using Pearson or Spearman’s rank correlation.
Results
From an FGR cohort originally including 51 fetuses, three infants died in the neonatal period, six had incomplete perinatal hemodynamic measurements, another two declined follow-up at perinatal inclusion, and one withdrew consent for the whole study. At the age of 4 years, three infants were lost to follow-up and the parents of 12 infants withdrew from follow-up after initial agreement. In three children, buccal DNA sampling was denied or not feasible due to severe developmental problems. This resulted in the inclusion of 21 children with complete information on the CPR, DNA methylation levels, and neurodevelopmental test results. Children lost to or declining follow-up and DNA sampling (n = 20) less often required intensive care, but did not significantly differ in gestational age, birth weight (z-score), head circumference (z-score), PI of the umbilical artery (not more often above the 95th percentile), presence or absence of fetal brain-sparing, postnatal cerebral oxygenation, or gestational and neonatal complications from the included study population (Supplementary Table S2).
Characteristics of the study population
Eight children had evidence of fetal brain-sparing, while 13 children did not. Impaired flow in the umbilical artery (i.e., decreased, absent, or reversed end-diastolic flow, implying greater placental resistance) at last prenatal ultrasound was significantly more often observed in children showing fetal brain-sparing (Table 2). Moreover, in the same children, placental weight tended to be more often below the 10th percentile. Placental signs of infection were infrequent and equally distributed among groups. Maternal and gestational characteristics were largely similar, but delivery mode and duration of pregnancy were significantly different. Children with fetal brain-sparing had to be delivered by cesarean section more frequently and were born at a younger gestational age. Before birth, infants delivered by Cesarean section more often experienced heart rate decelerations as detected by cardiotocography (8 vs. 0, p = 0.015) and impaired flow in the umbilical artery (12 vs. 2, p = 0.020). Spontaneous or induced labor before Cesarean delivery was only seen in four cases, of which one was preterm and another one belonged to the group with fetal brain-sparing. There were no differences in sex. After birth, infants with fetal brain-sparing had significantly higher regional cerebral oxygen saturations on day 2, which was also highly related to an abnormally increased blood flow in the middle cerebral artery (p = 0.037), but not impaired umbilical flow, gestational age, Cesarean delivery, or birth weight z-score (p > 0.1). At 4 years of age, infants with fetal brain-sparing had better total behavior and executive functions, in particular, better externalizing behavior and inhibitory self-control (i.e., lower T-scores). Two infants (15%) without fetal brain-sparing were reported to be diagnosed with or highly suspected of autism spectrum disorder. Neither of the two groups showed intraventricular hemorrhage nor periventricular ischemic lesions as assessed by postnatal cranial ultrasound.
Table 2. Cohort characteristics
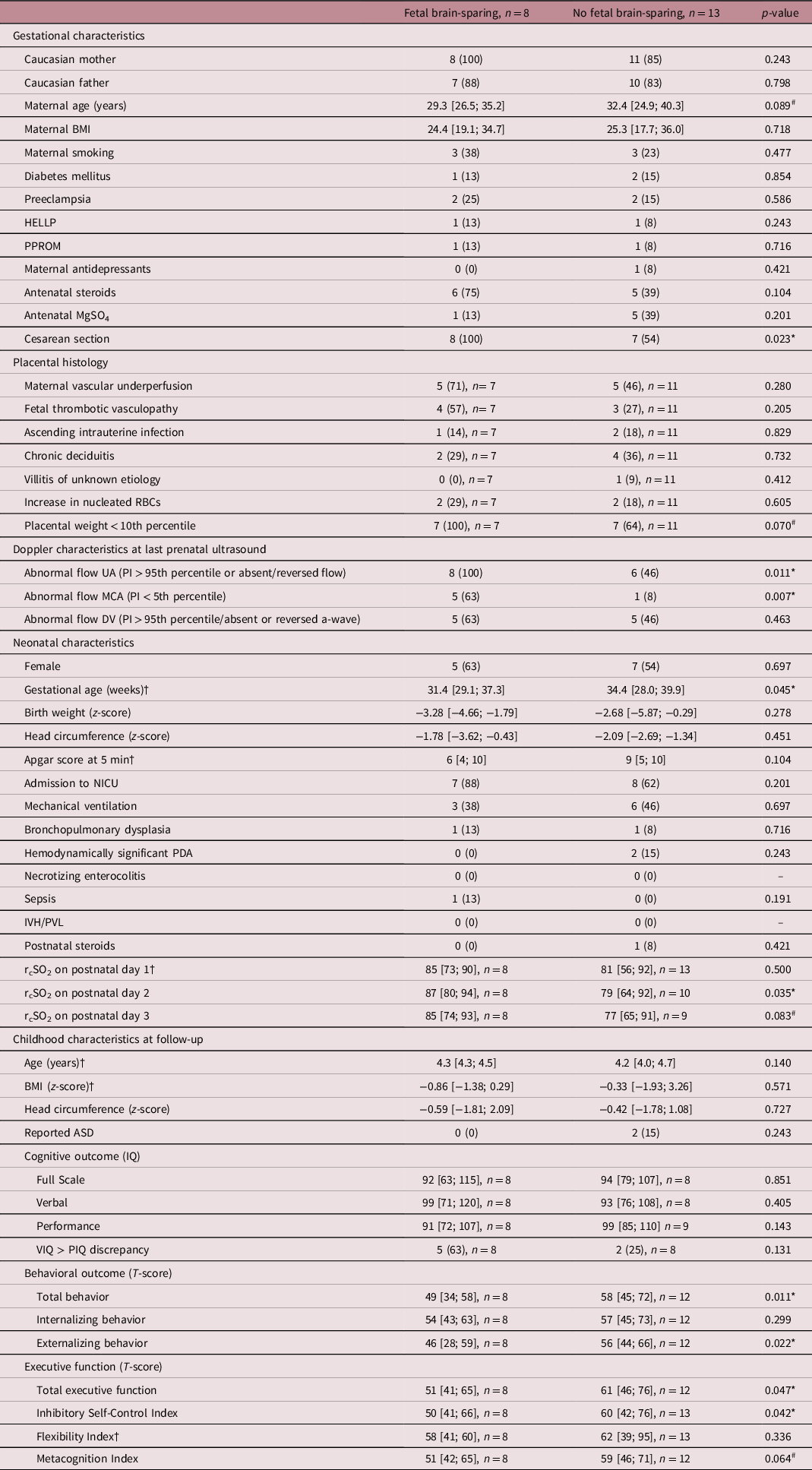
For good comparability, data are presented as mean/median [total range] or absolute number (percentage). †indicates, where a Mann–Whitney U test was used and a median is presented due to non-normality of the data. T-scores are to be interpreted as the lower, the better. # and * present differences between both groups at p < 0.1 and p < 0.05, respectively. ASD, autism spectrum disorder; BMI, body mass index; DV, ductus venosus; HELLP, syndrome of hemolysis, elevated liver enzymes, and low platelets; IQ, intelligence quotient; IVH, intraventricular hemorrhage; MCA, middle cerebral artery; MgSO4, magnesium sulfate; NICU, neonatal intensive care unit; PDA, patent ductus arteriosus; PI, pulsatility index; PIQ, performance intelligence quotient; PPROM, prolonged premature rupture of membranes (>12 h); PVL, periventricular leukomalacia; RBC, red blood cell; rcSO2, regional cerebral oxygen saturation (measured with near-infrared spectroscopy); UA, umbilical artery; VIQ, verbal intelligence quotient.
Differences in methylation between FGR children with and without brain-sparing
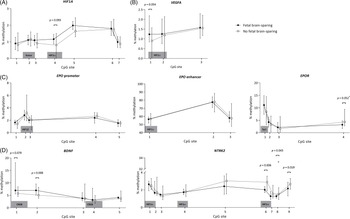
The methylation patterns for FGR children with and without brain-sparing are presented per analyzed genomic region in Fig. 2. In children with fetal brain-sparing, there was a trend toward a significantly higher percentage of methylation at CpG site 4 of the analyzed HIF1A locus than in FGR children without fetal brain-sparing (mean [μ] ± standard deviation [SD] of 1.16 ± 0.28 vs. 0.086 ± 0.40, p = 0.093), while all other CpGs were not methylated differently (Table 3). CpG 4 lies exactly within a binding site for HIF1α, as demonstrated in Table 1.
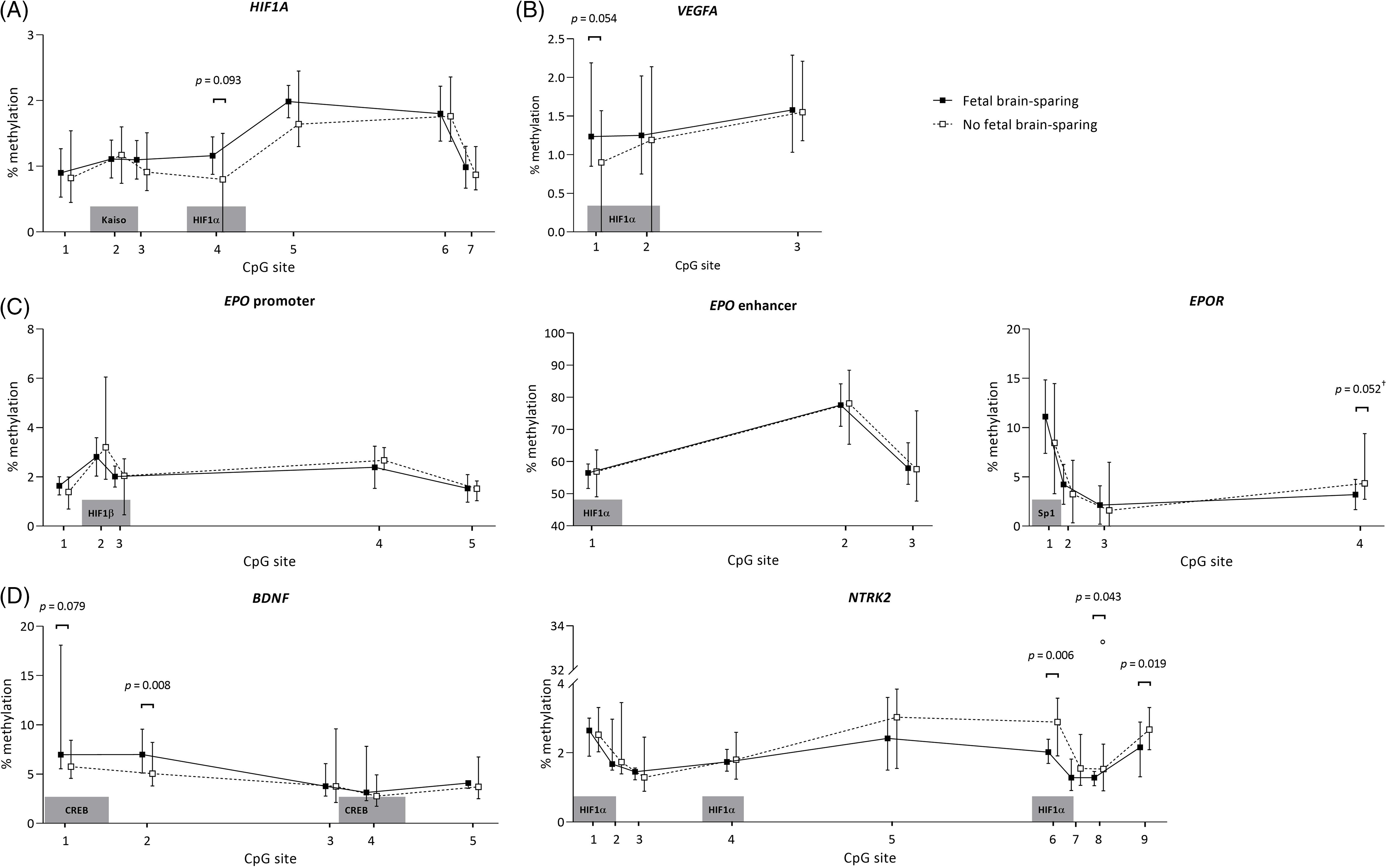
Fig. 2. Buccal DNA methylation patterns at selected regions of (A) HIF1A, (B) VEGFA, (C) EPO and EPOR, and (D) BDNF and its receptor gene NTRK2 in 4-year-old children born following fetal growth restriction with (n = 8) and without fetal brain-sparing (n = 13). Data are presented as medians (total range); the circle represents an extreme outlier (33.21%). The p-values given were gained using Student’s t-test or Mann–Whitney U test, involved numbers (n) per analyzed CpG, and group are given in Table 3. †p > 0.1 when adjusted for gestational age in multivariate regression analysis. BDNF, brain-derived neurotrophic factor; CpG, 5′-cytosine-phosphate-guanine-3′ dinucleotide; CREB, cyclic adenosine monophosphate response element binding protein; EPO, erythropoietin; EPOR, erythropoietin receptor; HIF1A/α, hypoxia-inducible factor-1 alpha; HIF1β, hypoxia-inducible factor-1 beta; NTRK2, neurotrophic tyrosine kinase, receptor, type 2; Sp1, specificity protein 1; VEGFA, vascular endothelial growth factor A.
Table 3. Differences in percentage methylation per CpG of selected gene locations at 4 years of age between children born following fetal growth restriction with or without brain-sparing, as assessed with Student’s t-test or Mann–Whitney U test
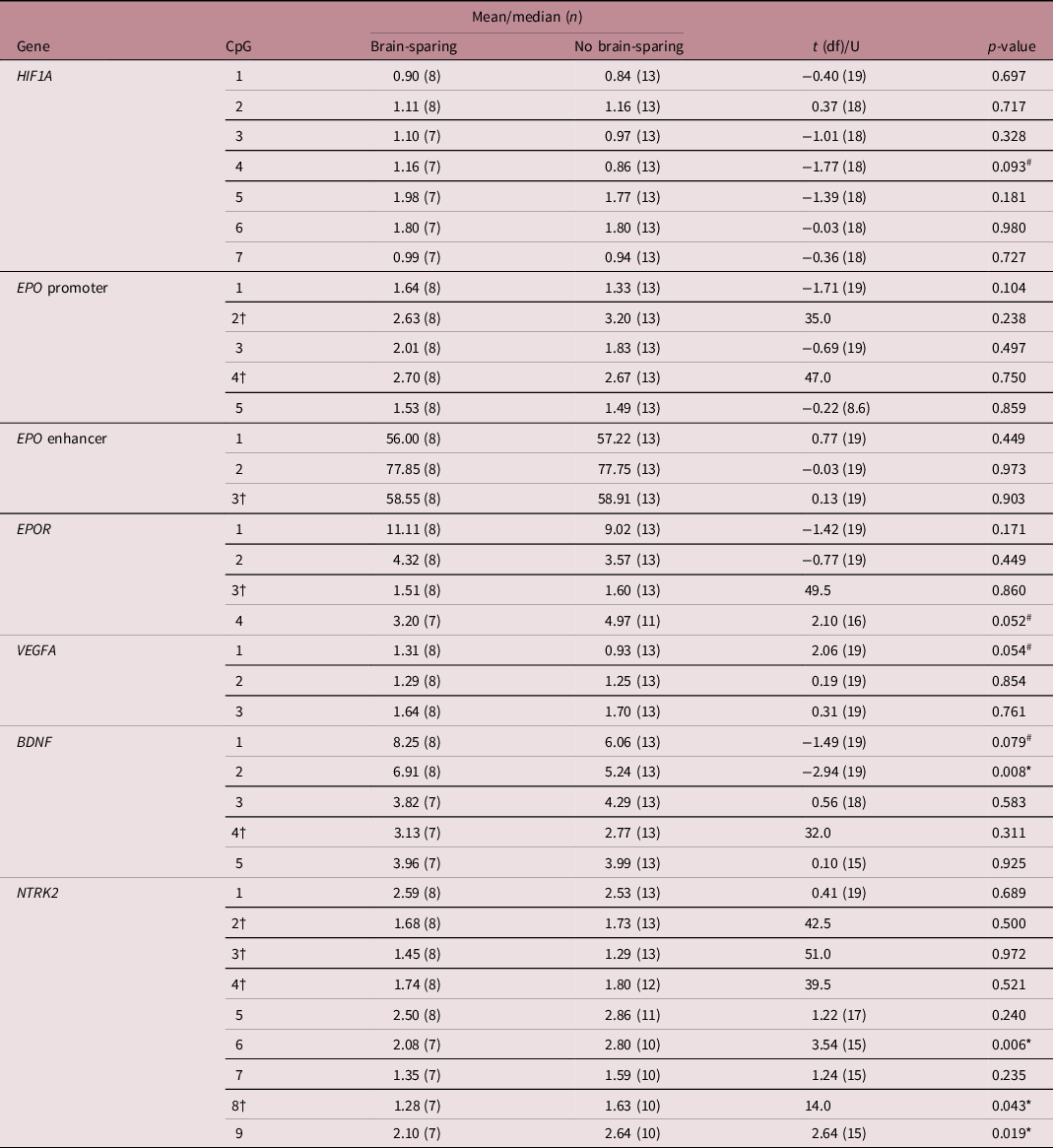
†Mann–Whitney U test was used due to non-normality of the data, for which median and U are given. # and * mark differences between means or medians at (an uncorrected) p < 0.1 and p < 0.05, respectively. BDNF, brain-derived neurotrophic factor; CpG, 5′-cytosine-phosphate-guanine-3′ dinucleotide; df, degrees of freedom; EPO, erythropoietin; EPOR, erythropoietin receptor; HIF1A, hypoxia-inducible factor-1 alpha; NTRK2, neurotrophic receptor tyrosine kinase 2; VEGFA, vascular endothelial growth factor A.
The analyzed EPO promoter and enhancer region were not methylated differently, but CpG position 4 of the EPOR gene tended to be significantly less methylated in FGR children with fetal brain-sparing than without (μ ± SD = 3.20 ± 1.53 vs. 4.97 ± 1.85, p = 0.052; Table 3). Methylation levels of CpGs 1–3 of EPOR were not different between the groups, and unfortunately, CpG 5–7 had to be excluded from analyses due to insufficient quality of measurements.
In children with fetal brain-sparing, we found a trend toward significantly higher methylation levels at CpG 1 of the selected VEGFA locus (μ ± SD = 1.31 ± 0.43 vs. 0.93 ± 0.40, p = 0.054; Table 3), which lies within an HRE (Fig. 2). Furthermore, FGR children with fetal brain-sparing had significantly higher methylation levels at CpG 2 (μ ± SD = 6.91 ± 1.41 vs. 5.24 ± 1.17, p = 0.008) and a tendency toward higher methylation of CpG 1 (μ ± SD = 8.25 ± 4.04 vs. 6.06 ± 1.15, p = 0.079) of the selected BDNF locus (Table 3). Only the latter lies within a transcriptionally important CREB binding site (Figs. 1 and 2), but methylation levels of both CpG sites highly correlated (Supplementary Table S3). The BDNF receptor gene NTRK2 demonstrated significantly lower methylation levels at CpG 6 (which lies within an HRE, μ ± SD = 2.08 ± 0.30 vs. 2.80 ± 0.54, p = 0.006), CpG 8 (median [interquartile range] = 1.28 [1.09; 1.44] vs. 1.63 [1.37; 2.13], p = 0.043), and CpG 9 (μ ± SD = 2.10 ± 0.51 vs. 2.64 ± 0.34, p = 0.019) in FGR children with fetal brain-sparing (Table 3; Fig. 2). Correlation coefficients between these CpG sites were high (Supplementary Table S2). CpG 8 of NTRK2 contained an extreme outlier within the group of FGR children without fetal brain-sparing (33.21% methylation). Excluding the outlier did not relevantly change our results (μ ± SD = 1.28 ± 1.58 vs. 1.62 ± 0.43, t(11) = 2.23, p = 0.049). As quality of methylation analysis was high, the presented data in Table 3 and Fig. 2 include this outlier.
Multiple regression analysis
Gestational age, which was significantly lower in children with fetal brain-sparing, also significantly correlated with methylation of EPOR CpG 4 (Pearson’s correlation coefficient = 0.479, p = 0.038). As gestational age is known to have a separate effect on DNA methylation, we included it as a confounder in a linear regression model. After correcting for gestational age, the association between brain-sparing and methylation of EPOR at CpG 4 lost its significance (B [95% confidence interval] = −1.31 [−3.19; 0.58], t = −1.48, p = 0.160).
Association between differentially methylated CpGs and neurodevelopmental outcome
To secondarily assess whether differential CpG methylation among children with and without brain-sparing may explain differences in neurodevelopmental outcome between the two groups, correlation coefficients between CpG methylation levels and outcome were calculated (Table 4). Increased methylation of CpG 4 of HIF1A was associated with a trend toward higher Full Scale IQ (Pearson coefficient = 0.449, p = 0.081) and higher Verbal IQ (Pearson coefficient = 0.429, p = 0.098). Moreover, methylation of CpG 1 of the VEGFA locus significantly and inversely correlated with Performance IQ (Pearson coefficient = −0.660, p = 0.003), while hypermethylation of CpG 2 (and to a minor extent CpG 1) of BDNF correlated with better executive function (i.e., lower T-scores), in particular, better inhibitory self-control (Pearson coefficient = −0.493, p = 0.020).
Table 4. Correlation coefficients between the percentage methylation of brain-sparing associated CpGs and neurodevelopmental outcome at 4 years following fetal growth restriction
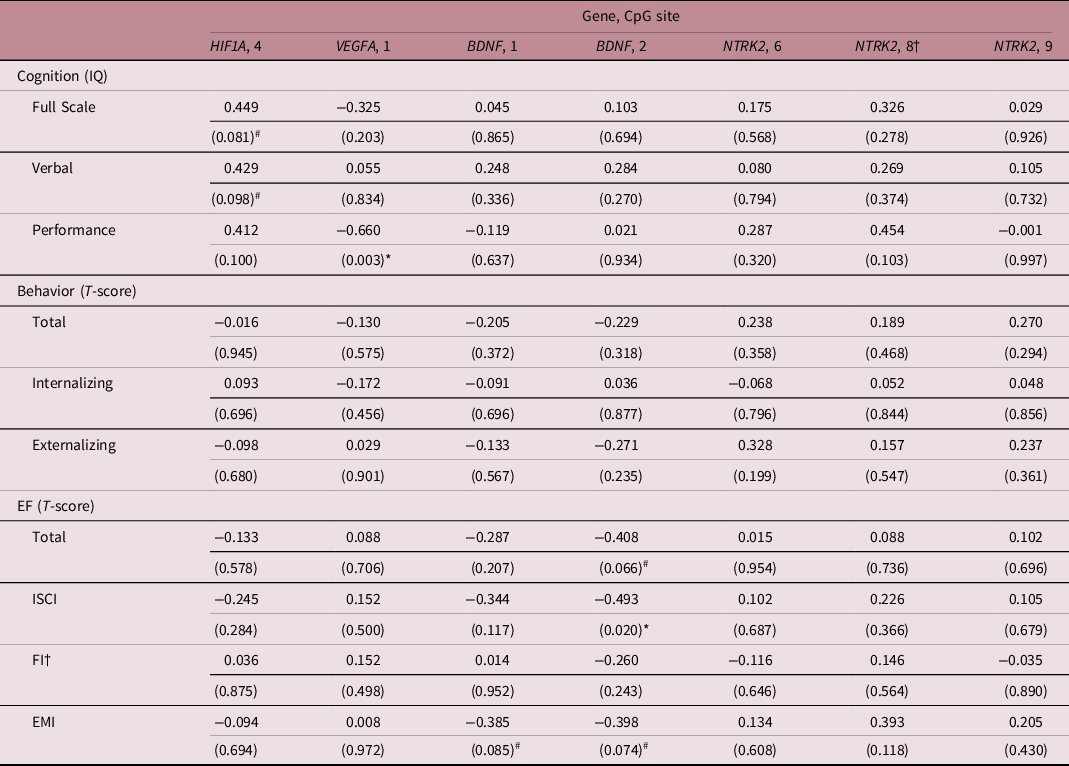
†Spearman’s rank correlation analysis was used due to non-normality of the data. P-values are given in brackets and were not corrected for multiple testing. # and * mark associations at p < 0.1 and p < 0.05, respectively. T-scores are to be interpreted as the lower, the better. BDNF, brain-derived neurotrophic factor; CpG, 5′-cytosine-phosphate-guanine-3′ dinucleotide; EF, executive function; EMI, Emergent Metacognition Index; EPOR, erythropoietin receptor; FI; Flexibility Index; HIF1A, hypoxia-inducible factor-1 alpha; ISCI; Inhibitory Self-Control Index; IQ, Intelligence Quotient; NTRK2, neurotrophic tyrosine kinase, receptor, type 2; VEGFA, vascular endothelial growth factor A.
Discussion
In this prospective follow-up study we compared the buccal DNA methylation status of predefined genomic regions of HIF1A and some of its neurotrophic target genes between FGR children with and without fetal brain-sparing, relating it to their neurodevelopmental outcome. The regions analyzed were selected based on their proven or theoretical potential to alter activity of the respective gene in an oxygen-dependent manner. We found that FGR children with prenatal evidence of brain-sparing showed a trend toward hypermethylation at the HRE of HIF1A and VEGFA. Moreover, we found hypermethylation at a CREB binding site within the promoter region of BDNF exon 4 and hypomethylation at an HRE located within the promoter region of its receptor NTRK2.
HIF1α regulates the cellular response to hypoxic conditions and among its targets are many neurotrophic factors. In a previous study, we found that preferential perfusion of the fetal brain in FGR was associated with better neurodevelopmental outcome at 4 years of age than FGR without fetal brain-sparing. Reference Richter, Salavati and Kooi5 This seemed to be mediated through higher cerebral oxygen saturations as suggested by postnatal tissue oxygenation monitoring with near-infrared spectroscopy. In the same cohort, we now find a trend toward hypermethylation at the autoregulatory HRE of the HIF1A promoter in the buccal DNA of these children. Since buccal methylation patterns closely correlate with those of neuronal DNA due to their common ectodermal origin, this may reflect suppression of the hypoxic response in brain tissue. Reference Smith, Kilaru and Klengel14 This could be caused by higher cerebral oxygen saturations, which are evident in FGR neonates with fetal brain-sparing. Reference Tanis, Boelen and Schmitz4 Moreover, hypermethylation of this locus was associated with a trend toward better Full Scale and Verbal IQ. This may be mediated through its effects on neurotrophic factors, since postnatal cranial ultrasounds did not demonstrate a difference in ischemic lesions between FGR infants with and without fetal brain-sparing. However, it needs to be emphasized at this point that these trend may also merely represent accidental findings and require confirmation.
Several genetic factors play critical neurotrophic roles during early human brain development, whose expression has shown to be affected by hypoxia. VEGFA expression is increased by HIF1α with important pro-angiogenic effects under hypoxic conditions but also stimulatory effects on axonal outgrowth and the proliferation, migration, and survival of neurons and neuroglia. Reference Carmeliet and de Almodovar46 Oosthuyse et al. demonstrated impaired hypoxic upregulation of neural VEGF and motor neuron degeneration in knockout mice lacking another HRE in the VEGFA promoter. Reference Oosthuyse, Moons and Storkebaum47 Since hyperoxia downregulates VEGF expression, significantly higher postnatal cerebral oxygenation levels in FGR children with fetal brain-sparing than in those without brain-sparing may possibly explain the observed trend toward hypermethylation at the HRE of VEGFA in FGR children with fetal brain-sparing. Although this tendency toward increased methylation at this locus may also be an accidental finding, if found to be true and related to reduced expression of VEGFA, it could explain the inverse correlation between VEGFA methylation and Performance IQ at 4 years, since Performance IQ is related to motor function. Reference Kopp, Beckung and Gillberg48 That there may be a link between early cerebral oxygenation levels, altered VEGFA levels and neurodevelopmental functioning, was already suggested by us in a previous study, which demonstrated a lower Performance IQ in children with comparably high oxygen saturations during the first days after birth. Reference Richter, Salavati and Kooi5
While above theory is based on a small cohort and studies testing the methylation of the analyzed HRE in relation to expression of VEGFA are not yet available, it further remains debatable whether increased buccal VEGFA methylation at later age could indeed result from fetal brain-sparing. We recently demonstrated an association between fetal brain-sparing and hypermethylation of the same CpG of VEGFA in the placental tissue of this FGR cohort. Reference Bekkering, Leeuwerke and Tanis42 However, one may rather expect placental hypomethylation at this locus, since fetal brain-sparing is generally accepted to be a compensatory fetal response to placental hypoxia. Instead, both placental and buccal hypermethylation may therefore reflect an anti-angiogenic state during early fetoplacental development, which leads to placental insufficiency and subsequently fetal brain-sparing. Reference Helmo, Lopes and Carneiro49 This angiogenic dysbalance seems to persist in the neonate. Reference Hentges, Silveira and Procianoy50 More severe placental insufficiency in children with fetal brain-sparing, which may be suggested by a decreased flow in the umbilical artery (indicating a higher placental resistance) and a tendency towards lower placental weight, could therefore also explain our findings. Although an angiogenic dysbalance (lower circulating VEGF levels) has also been evidenced in patients with ASD, we have reported in our previous paper that both children also presented with impaired umbilical flow even if no signs of fetal brain-sparing were present. Reference Richter, Salavati and Kooi5,Reference Emanuele, Orsi, Barale, di Nemi, Bertona and Politi51 Regardless of its origin, VEGFA hypermethylation does not seem to benefit neurodevelopmental outcome.
Although the BDNF gene does not contain any HREs, transcriptional activation upon hypoxia has shown to occur through interaction of the transcription factor CREB with the promoter region of exon 4, mediated through EPO-enhanced phosphorylation of CREB. Reference Viviani, Bartesaghi and Corsini39 Accordingly, we found significant hypermethylation close to the designated binding sites of CREB at this promoter in FGR children with fetal brain-sparing. However, we did not observe any significant differences in methylation of the selected HRE regions in EPO, suggesting that this may not fully explain altered BDNF methylation. Moreover, hypermethylation of BDNF seemed associated with better executive functioning, in particular significantly better inhibitory self-control, although we expected hypermethylation to cause poorer executive functioning by reducing expression of BDNF. However, the analyzed CREB binding sites have also been implicated in calcium-mediated, activity-dependent upregulation of BDNF through N-methyl-D-aspartate (NMDA)-receptor activation by glutamate. Reference Fang, Chartier and Sodja38,Reference Kundakovic, Gudsnuk, Herbstman, Tang, Perera and Champagne41 BDNF is known to sustain NMDA activation through TrkB signaling, creating a positive feedback loop, which promotes neuronal sprouting and synaptogenesis but may also lead to hyperexcitability. Reference Murray and Holmes52 Our findings may therefore reflect a reduction of perinatal hypoxia-induced glutamate excitotoxicity through hypermethylation of BDNF in the presence of fetal brain-sparing. Reference Burd, Welling, Kannan and Johnston53 This may also contribute to increased BDNF levels in autism spectrum disorder, which has been related to an excitatory/inhibitory imbalance and, in our cohort, was reported in 15% of FGR children without fetal brain-sparing. Reference Qin, Feng, Cao, Wu, Loh and Cheng54,Reference Gao and Penzes55
Interestingly, regarding the receptors of neurotrophic factors, brain-sparing was also significantly associated with hypomethylation of the BDNF receptor gene NTRK2 at CpG position 6, which corresponds to HRE1 reported by Martens et al., and the closely located CpG positions 8 and 9. Reference Martens, Kirschner, Warnecke and Scholz11 Likewise, we found a trend toward significant hypomethylation of EPOR, although this seemed to be confounded by lower gestational age, supporting the hypothesis that methylation of this region is unaffected by oxygenation status but involved in developmental downregulation of EPOR. Reference Wallach, Zhang and Hartmann35 Hypomethylation of the HRE of NTRK2, however, was unexpected and may be related to placental hypoxia rather than brain-sparing, since it also did not correlate with neurodevelopmental outcome. This is in line with studies demonstrating elevated NTRK2 levels in the placental tissue of FGR pregnancies to stimulate endothelial cell survival and angiogenesis. Reference Dunk, Roggensack and Cox56 Although low BDNF levels have also been demonstrated in early preeclampsia and may contribute to an anti-angiogenic placental environment, the analyzed promoter region of exon 4 is highly tissue-specific for the brain, which may also explain paradoxical differences in methylation between BDNF and its receptor gene. Reference Boulle, Van Den Hove and Jakob57,Reference D’Souza, Kilari, Joshi, Mehendale, Pisal and Joshi58
To our knowledge, this is the first study analyzing the association between fetal brain-sparing and buccal DNA methylation patterns of neurodevelopmentally important genes in primary school children born following FGR. The strength of our study lies within the specific analyses of binding sites implicated in the hypoxic upregulation of these genes and their relationship to outcome established by prospective neurodevelopmental testing. However, we acknowledge some limitations. First, the sample size of this study was small, which limited our power to detect significant associations. In addition, it limited our study to a candidate gene approach, while an epigenome-wide association study would yield far more information. Second, we performed multiple testing without controlling for it, as we considered this a hypothesis-generating study. However, while this reduced type II error, it may also have increased our false discovery rate. Reported trends (p < 0.1), in particular, but also significant findings (p < 0.05) are therefore to be interpreted with great caution and require validation by larger studies. Third, due to the small sample size, we may have neglected some important confounders of the association between brain-sparing and methylation as well as the association between methylation and neurodevelopmental outcome, such as sex or ethnicity. Moreover, although we tried to study causation between brain-sparing and altered methylation patterns by analyzing oxygen-dependent regulatory genomic regions, some of our findings may also be explained by underlying anti-angiogenic pathology of placental insufficiency or resulting hypoxia, which could also explain differences in DNA methylation. Reference Weber, Hellmann and Stadler9 Finally, differences in methylation were small, and it remains to be investigated whether they would be similar in neuronal DNA and sufficient to alter gene expression. Therefore, we encourage replication of our findings by larger trials with additional gene expression analysis in both buccal and neuronal DNA.
In conclusion, this explorative study, limited in sample size, showed that fetal brain-sparing in FGR is associated with a trend toward buccal hypermethylation of HIF1A and VEGFA, significant hypermethylation of BDNF, and significant hypomethylation of NTRK2 at regions implicated in oxygen-mediated regulation of these genes. Moreover, increased methylation of these regions within BDNF and VEGFA was significantly associated with better inhibitory self-control and poorer Performance IQ, respectively. However, before we can draw conclusions about possible causation and mechanisms behind these findings, they need to be confirmed by larger hypothesis-testing studies, which also need to involve gene expression analysis.
Supplementary Material
To view supplementary material for this article, please visit https://doi.org/10.1017/S2040174421000374
Acknowledgements
We thank A. Britt Foreman and Sahar Salavati for their help and assistance during follow-up and Anne den Heijer for her expertise, advice, and training during neurodevelopmental testing. We thank Mirthe H. Schoots for analyzing the placentas of this FGR cohort, thereby providing valuable information to this work.
Financial Support
This study was part of the research program of the Research Institute of Behavioral and Cognitive Neurosciences (BCN), Graduate School of Medical Sciences, University of Groningen, participation in which is financially supported by the Junior Scientific Master Class of the University Medical Center Groningen, University of Groningen, The Netherlands.
Conflicts of Interest
None.
Ethical standards
The authors assert that all procedures contributing to this work comply with the ethical standards of the relevant national guidelines on human experimentation (Wet medisch-wetenschappelijk onderzoek met mensen) and with the Helsinki Declaration of 1975, as revised in 2008, and has been approved by the institutional committees (Medisch Ethische Toetsingscommissie van het UMCG).