Introduction
The genus Diastolaimus Rahm, 1928 includes six species (Andrássy, Reference Andrássy and Franz1984; Cid del Prado, Reference Cid del Prado2012), most of them poorly described. The main morphological diagnostic characters of the genus are the presence of seta-like inner labial papillae, structure and proportion of the cheilostom–gymnostom, didelphic-amphidelphic female genital system, male and female tails with elongate mucro having bifurcate tip, and the morphology of the spicules and the gubernaculum. However, in general, some of these diagnostic characters remain unknown or were described inaccurately.
Diastolaimus grossus (Truskova & Eroshenko, 1977) Andrássy, 1984, was originally described as Fescia grossa by Truskova & Eroshenko (Reference Truskova and Eroshenko1977) from Chuguyevsky district, Primorsky Krai (Far Eastern Siberia, Russia) examining four females and two males collected from the branches of Abies sp. In the present paper, D. grossus is morphologically, morphometrically and molecularly characterized through material found in two forest locations in the Czech Republic and Bosnia and Herzegovina. In addition, new molecular data of D. grossus (18S ribosomal DNA (rDNA): OM691517; 28S rDNA: OM691516), Macrolaimus canadensis Sanwal, 1960 (18S rDNA: OM691513; 28S rDNA: OM691511, OM691512) and Macrolaimus ruehmi Andrássy, 1966 (18S rDNA: OM691506; 28S rDNA: OM691514, OM691515) from the Czech Republic, Macrolaimus arboreus Truskova & Eroshenko, 1977 from Iran (28S rDNA: MF996701, MF996702) and Macrolaimus crucis Maupas, 1900 (18S rDNA: OM691510; 28S rDNA: OM691509) from Spain are provided.
Materials and methods
Sampling and nematode extraction
Specimens of D. grossus were extracted from the bark or barked wood samples taken from Náměšť na Hané, Czech Republic (Salix sp.) and from Banja Luka, Bosnia and Herzegovina (Pinus nigra Arnold). Molecular data of M. canadensis and M. ruehmi were obtained from samples taken during the monitoring of Macrolaimus species (Abolafia et al., Reference Abolafia, Ruiz-Cuenca, Foit and Čermák2018). Nematodes from the collected samples were extracted using a modified Baermann (Reference Baermann1917) funnel technique, killed by heat, fixed in 4% formalin, transferred to pure glycerine following De Grisse's (Reference De Grisse1969) technique for the Czech and Iranian specimens and following Siddiqi's (Reference Siddiqi1964) technique for the Spanish specimens, and mounted on permanent glass slides according to the de Maeseneer & d'Herde (Reference de Maeseneer and d'Herde1963) technique, but somewhat modified.
Light microscopy (LM)
Measurements were taken for specimens mounted on permanent slides. Pictures were taken with a Leica DM2500 (Leica, Wetzlar, Germany) light microscope provided with differential interference contrast optics and a Leica DMC2900 camera. Micrographs were combined using Adobe® Photoshop® CS (Adobe Inc., San José, USA). The terminology used for the morphology of stoma and spicules follows the proposals by De Ley et al. (Reference De Ley, van de Velde, Mounport, Baujard and Coomans1995) and Abolafia & Peña-Santiago (Reference Abolafia and Peña-Santiago2017), respectively.
Scanning electron microscopy (SEM)
Specimens preserved in glycerine were selected for observation under SEM according to Abolafia (Reference Abolafia2015). The nematodes were hydrated in distilled water, dehydrated in a graded ethanol-acetone series, critical-point dried, coated with gold and females observed with a Zeiss Merlin microscope (5 kV) (Zeiss, Oberkochen, Germany) and males with a Hitachi SU 8010 (Hitachi, Tokyo, Japan).
DNA amplification and sequencing
Czech specimens
DNA amplification was performed by heat gently inactivated specimens identified by LM. DNA was extracted as follows: single specimens were homogenized in 50 μl of nematode lysis buffer (10 mm Tris-hydrochloride (Tris-HCl), pH 8.8; 1 mm ethylene-diamine-tetraacetic acid (EDTA); 1% Triton X-100 (v/v); 100 μg ml−1 proteinase K) in a 1.5 mL Eppendorf tube using a micropestle. Sample incubation at 55°C for 1 h and subsequently at 95°C for 10 min followed. The resulting DNA extract was used as a template for polymerase chain reaction (PCR). Amplification was performed using GoTaq® G2 Flexi DNA Polymerase (Promega, Madison, Wisconsin, USA). The primers used for amplification of the region of 18S ribosomal RNA (rRNA) gene were the forward primers 988 F (5′-CTCAAAGATTAAGCCATGC-3′) and 1813F (5′-CTGCGTGAGAGGTGAAAT-3′) and the reverse primers 1912R (5′-TTTACGGTCAGAACTAGGG-3′) and 2646R (5′-GCTACCTTGTTACGACTTTT-3′) (Holterman et al., Reference Holterman, van der Wurff, van den Elsen, van Megen, Bongers, Holovachov, Bakke and Helder2006). The primers used for amplification of the D2–D3 region of 28S rRNA gene were forward primer D2A (5′-ACAAGTACCGTGAGGGAAAGTTG-3′) and the reverse primer D3B (5′-TCGGAAGGAACCAGCTACTA-3′) (synthetized by Integrated DNA Technologies) targeting the D2–D3 region of 28S rRNA gene (Nunn, Reference Nunn1992; De Ley et al., Reference De Ley, Felix, Frisse, Nadler, Sternberg and Thomas1999). The total volume of the reaction mix was 25 μl containing 1× Colorless GoTaq® Flexi Buffer (Promega, Madison, Wisconsin, USA); 1.25 mm magnesium chloride (MgCl2); 250 nM deoxyrionucleoside triphosphates; 600 nM of each primer; 1.25 U of GoTaq® G2 Flexi DNA Polymerase (Promega, Madison, Wisconsin, USA) and 2 μl of undiluted DNA extract. The reaction was performed in the XP thermal cycler (Bioer Technology, Hangzhou, China) and consisted of 3 min at 94°C initial denaturation followed by 40 cycles of 30 s at 94°C, 1 min at 55°C and 2 min at 72°C, with a final elongation step of 7 min at 72°C. Ten microlitres of the PCR product were run on 1.5% Tris-Borate-EDTA (TBE)-buffered agarose gel to check the amplification. The size of PCR product was approximately 800 bp. The remaining 15 μl of PCR product was submitted for sequencing to the Centre of Region Haná for Biotechnological and Agricultural Research, Institute of Experimental Botany (Olomouc, Czech Republic). Obtained DNA sequences were analysed using the Geneious Bioinformatics software platform (Biomatters, Auckland, New Zealand).
Spanish specimens
Nematode DNA was extracted from single fresh individuals using the proteinase K protocol and PCR assays as described by Castillo et al. (Reference Castillo, Vovlas, Subbotin and Troccoli2003), but somewhat modified (Archidona-Yuste et al., Reference Archidona-Yuste, Navas-Cortés, Cantalapiedra-Navarrete, Palomares-Rius and Castillo2016). Specimens were cut into small pieces using a sterilized dental anaesthesia needle on a clean slide with 18 ml of Tris-EDTA buffer (10 mm Tris-HCl + 0.5 mm EDTA; pH 9.0), transferred to a microtube, adding 2 μl proteinase K (700 μg/ml−1) (Roche, Basel, Switzerland), and stored at −80°C within 15 min (for several days). The microtubes were incubated at 65°C (1 h), then at 95°C (15 min). For DNA amplification, 3 μl of the extracted DNA was transferred to a microtube containing the following: 0.6 μl of each primer (10 mm), 3 μl Master Mix Taq DNA Polymerase (5× Hot FirePol Blend Master Mix, Solis BioDyne, Tartu, Estonia) and double-distilled water (ddH2O) to a final volume of 20 μl. The primers used for amplification of the region of 18S rRNA gene were the forward primer 988F (5′-CTCAAAGATTAAGCCATGC-3′) and the reverse primer 1912R (5′-TTTACGGTCAGAACTAGGG-3′) (Holterman et al., Reference Holterman, van der Wurff, van den Elsen, van Megen, Bongers, Holovachov, Bakke and Helder2006). The primers used for amplification of the D2–D3 region of 28S rRNA gene were the forward primer D2A (5′-ACAAGTACCGTGAGGGAAAGTTG-3′) and the reverse primer D3B (5′-TCGGAAGGAACCAGCTACTA-3′). PCR cycle conditions were as follows: one cycle of 94°C for 15 min, followed by 35 cycles of 94°C for 45 s + annealing temperature of 55°C for 45 s + 72°C for 45 s, and finally one cycle of 72°C for 5 min. After DNA amplification, 5 μl of product was loaded on a 1% agarose gel in 0.5% Tris-acetate-EDTA (40 mm Tris, 20 mm glacial acetic acid and 2 mm EDTA; pH = 8) to verify the amplification using an electrophoresis system (Labnet Gel XL Ultra V–2, Progen Scientific, London, UK). The bands were stained with RedSafe (Intron Biotechnolog, Seongnam, South Korea) (20,000×) previously added to the agarose gel solution. The sequencing reactions of the PCR products were performed at Sistemas Genómicos (Paterna, Valencia, Spain) according to the Sanger et al. (Reference Sanger, Nicklen and Coulson1977) method.
Iranian specimens
DNA was extracted from nematodes using the method according to Holovachov et al. (Reference Holovachov, Boström, De Ley I, Nadler and De Ley2009). Specimens were picked using a fine-tipped needle and transferred to a 1.5 ml microtube containing 25 μl of ddH2O. The tube containing the nematode was crushed by a sterile needle. Following this procedure, 20 μl of Chelex 100 and 2 μl proteinase K (20 mg/ml) were added to the nematode substrate and the homogenate incubated at 56°C for 2 h and then at 95°C for 10 min. The supernatant was then extracted from the tube and stored at −20°C. The primers used for amplification of the D2–D3 region of 28S rRNA gene were the D2A (5′-ACAAGTACCGTGAGGGAAAGTTG-3′) and the D3B (5′-TCGGAAGGAACCAGCTACTA-3′) primers (Nunn, Reference Nunn1992; De Ley et al., Reference De Ley, Felix, Frisse, Nadler, Sternberg and Thomas1999). Subsequently, PCR was conducted with 8 μl of the PCR product of the nematode specimens, to which 2.5 μl of PCR buffer, 0.5 μl of DNTP, 1 μl of MgCl2, 0.3 μl of Taq-polymerase (CinnaGen, Iran), 1 μl of each primer listed above (10 pmol μl−1) and finally ddH2O were added, comprising a final volume of 25 μl. The amplification was carried out using an Eppendorf Mastercycler gradient (Eppendorf, Hamburg, Germany), with the following programme: initial denaturation for 3 min at 94°C, 37 cycles of denaturation for 45 s at 94°C, extension for 45 s at 56°C and annealing for 1 min at 72°C, and finally an extension cycle of 6 min at 72°C followed by a holding temperature of 4°C. After DNA amplification, 5 μl of product was loaded on a 2% agarose gel in TBE buffer (40 mm Tris, 40 mm boric acid and 1 mm EDTA) for evaluation of the DNA bands. The bands were stained with 50 mm ethidium bromide and visualized and photographed on an ultraviolet transilluminator. The PCR product was purified for sequencing by the Macrogen Corporation (Republic of Korea).
Phylogenetic analyses
For phylogenetic relationships, analyses were based on 18S rDNA and 28S rDNA fragments. Other segments of 18S rRNA and 28S rRNA gene sequences available in GenBank were aligned using ClustalX alignment in the program MEGA7 (Kumar et al., Reference Kumar, Stecher and Tamura2016). The sequence dataset was analysed with Bayesian inference using MrBayes 3.2.6 (Huelsenbeck & Ronquist, Reference Huelsenbeck and Ronquist2001; Ronquist & Huelsenbeck, Reference Ronquist and Huelsenbeck2003). The analysis under the GTR + G model was initiated with a random starting tree and run with the Markov chain Monte Carlo for 106 generations. Teratocephalus lirellus (AF036607 for the 18S phylogenetic tree and AB477073 for the 28S phylogenetic tree) was used as the outgroup. The phylogenetic trees were visualized and saved with the program FigTree 1.4.3 (Rambaut, Reference Rambaut2014).
Gower General Similarity coefficient of morphological characters
To examine the similarity of particular taxa of the family Chambersiellidae, a cluster analysis was implemented. The analysis was based on morphological diagnostic characteristics of particular populations, which were obtained within the present study or adopted from previously published studies: M_a-Ru: Macrolaimus arboreus (Russia; Truskova & Eroshenko, 1977); M_a-Ke: Macrolaimus arboreus (Iran; Shokoohi et al., 2018); M_ca-Ge: Macrolaimus canadensis (Germany; Fuchs (1938) as Macrolaimus crucis); M_ca-US/M_ca-Ca: Macrolaimus canadensis (USA/Canada; Sanwal, 1960); M_ca-CZ/M_ca-Co: Macrolaimus canadensis (Czech Republic/France; Abolafia et al., 2018); M_cu-Al: Macrolaimus crucis (Algeria; Maupas, 1900); M_cu-Sp: Macrolaimus crucis (Spain; Abolafia & Peña-Santiago, 2014); M_ri-Sa: Macrolaimus richteri Swart & Heyns, 1992 (South Africa); M_h-US: Macrolaimus hamatus Thorne, 1937 (USA); M_n_Ba: Macrolaimus natator Timm, 1960 (Pakistan); M_ru_Ge: Macrolaimus ruehmi (Germany; Rühm (1956) as M. crucis); M_ru-CZ/M_ru-BiH: Macrolaimus ruehmi (Czech Republic/Bosnia and Herzegovina; Abolafia et al., 2018); M_so-It: Macrolaimus somniorum Andrássy, 1984 (Italy); M_ta-US: Macrolaimus taurus Thorne, 1937 (USA); D_c-US: Diastolaimus croca (Massey, 1963) Andrássy, 1984 (USA; as Santafea croca Massey, 1963); D_d-US: Diastolaimus damalis Andrássy, 1984 (USA; as Santafea damalis Massey, 1966); D_g-Ru: Diastolaimus grossus Andrássy, 1984 (Russia; as Fescia grossa Truskova & Eroshenko, 1977); D_g-CZ/D_g-BiH: Diastolaimus grossus (Czech Republic/Bosnia and Herzegovina; present paper); D_m-Me: Diastolaimus mexicanus Cid del Prado, 2012 (Mexico); D_p-Br: Diastolaimus papillatus Rahm, 1928 (Brazil); Co_f-Ru: Cornilaimus furcillus Truskova & Eroshenko, 1977 (Russia); G_b-US: Geraldius bakeri (Sanwal, 1957) Sanwal, 1971 (USA; as Chambersiella bakeri Sanwal, 1957); G_g-Ec: Geraldius galapagoensis Cid del Prado, 2012 (Ecuador); Ch_r-US: Chambersiella rodens Cobb, 1920 (USA). Both, quantitative (such as morphometric) traits as well as qualitative traits were used for the analysis. For the purposes of quantitative traits, mean values for given population were used. Analysis was carried out separately for males and females because of many sex-associated morphological traits. Prior to the cluster analysis, Gower's dissimilarity coefficients were computed using the function ‘daisy’ in package ‘cluster’ in R 3.4.1 software, which was appropriate for the mixed-data (numeric/categorical) dataset that we had (Gower, Reference Gower1971). Subsequently, a matrix of Gower's dissimilarity coefficients was entered into hierarchical cluster analysis computed in R package ‘cluster’ using function ‘agnes’. Ward's (Reference Ward1963) clustering method was used because it exhibited higher agglomerative coefficients (0.9) than other available clustering methods.
Results
Diastolaimus grossus (Truskova & Eroshenko, 1977) Andrássy, 1984
Material from the Czech Republic
Nine females and three males in acceptable conditions. For morphometrics, see table 1.
Table 1. Morphometrics of D. grossus (Truskova & Eroshenko, 1977) Andrássy, 1984. Measurements in μm and in the form: mean ± standard deviation (range) where appropriate.
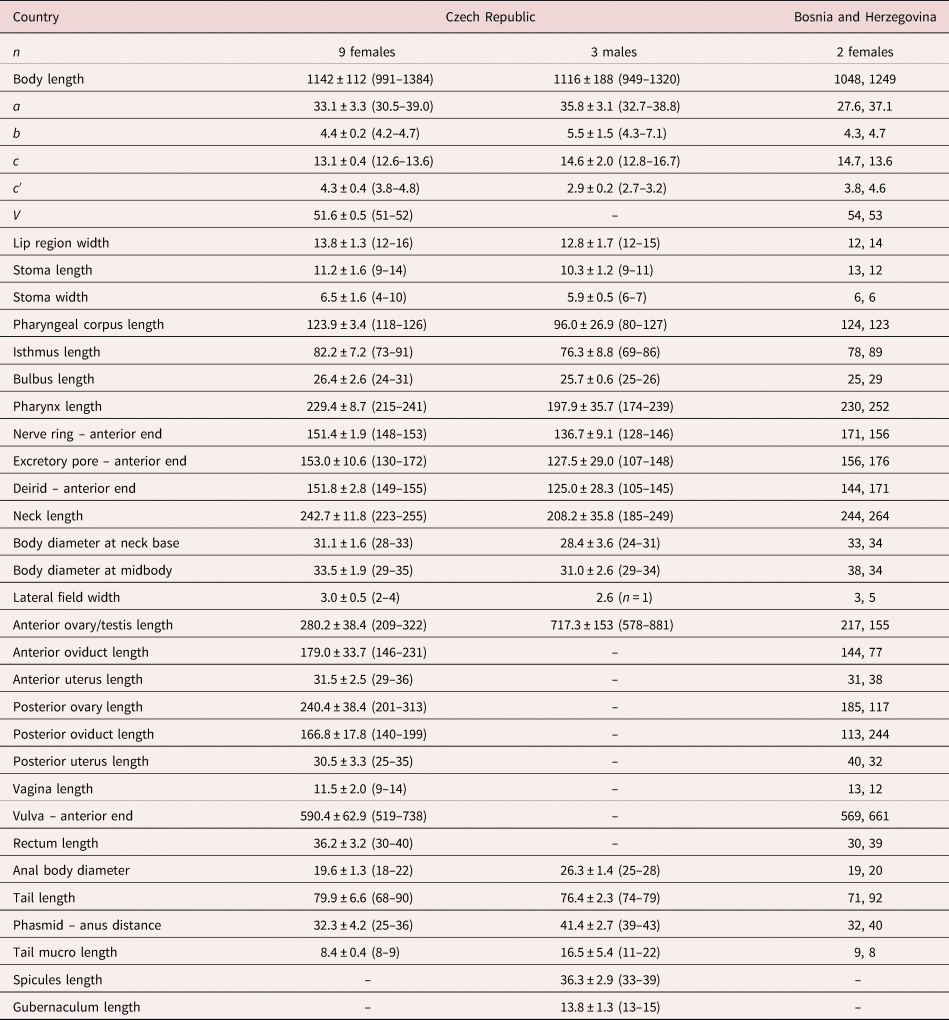
a = body length/body diameter; b = body length/pharynx length; c = body length/tail length; c′ = tail length/anal body diameter; V = (distance from anterior region to vulva/body length)x100.
Description
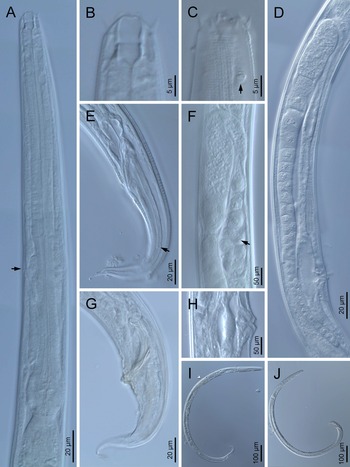
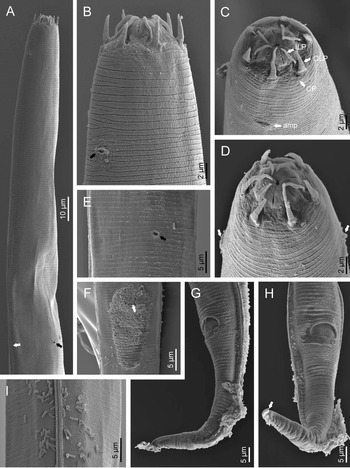
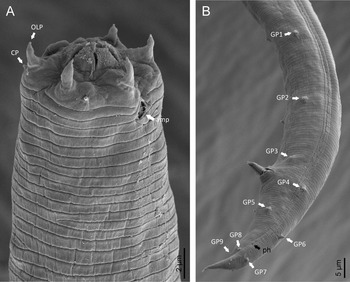
Adult (figs 1–3). Moderately slender nematodes of small size, 0.94–1.38 mm long. Body cylindrical, tapering towards both extremities, but more so towards the posterior end because the tail is conical. Habitus slightly curved ventrad after fixation, to an open ‘C’ shape. Cuticle ca. 1 μm thick, with very fine annuli, c. 1 μm wide, frequently appearing divided somewhere in its outline, hardly perceptible under LM but readily visible under SEM. Lateral fields 2–4 μm wide, consisting of two lines (three incisures) without transverse striation or any kind of areolation, starting at level of anterior part of corpus, and ending at tail tip. Lip region convex, continuous with the adjacent body, with the oral region strongly elevated or protruding. SEM observations: lips partially fused, but their inner (perioral) region, hemispheroid, forming six curved, triangular liplets covering the oral aperture; anterior sensilla arranged in three circles: six thin seta-like inner labial sensilla located at base of each liplet, six elongate-conoid outer labial sensilla and four papilliform cephalic sensilla. Amphidial apertures small, oval, located c. 15 annuli posteriorly to lips. Stoma 0.9–1.1 times longer than wide and 1.0 times as long as lip region diameter, consciously sclerotized, subdivided into cheilo-, gymno- and stegostom; cheilostom c. 50% of the stoma length, having cheilorhabdia consciously sclerotized, convergent (curved) and thinner anteriorly; gymnostom short, approximately one half of the cheilostom length, with arched gymnorhabdia arranged in three rows (examined under the LM); stegostom one half of the cheilostom length, with shallow funnel-shaped lumen, enveloped by the anterior end of pharynx, lacking visible rhabdia; dorsal pharyngeal gland opening at base of stegostom. Pharynx cephaloboid: pharyngeal corpus cylindrical, 1.3–1.6 times longer than isthmus, with procorpus and metacorpus similar in width; isthmus narrower than, and clearly delimited from corpus; basal bulb nearly pyriform, with strongly developed valvular apparatus located at its posterior half. Cardia short and subcylindrical 8.3 μm long in average. Nerve ring at 61–66% of neck length, encircling pharynx at level of anterior half of isthmus. Secretory–excretory gland cell located ventrally to isthmus, its excretory pore opening posterior to nerve ring, at 61–63% of neck length, at level of isthmus and nerve ring. Deirids located posterior to nerve ring, inside the lateral field, at 60–63% of neck length close and posterior to excretory pore.
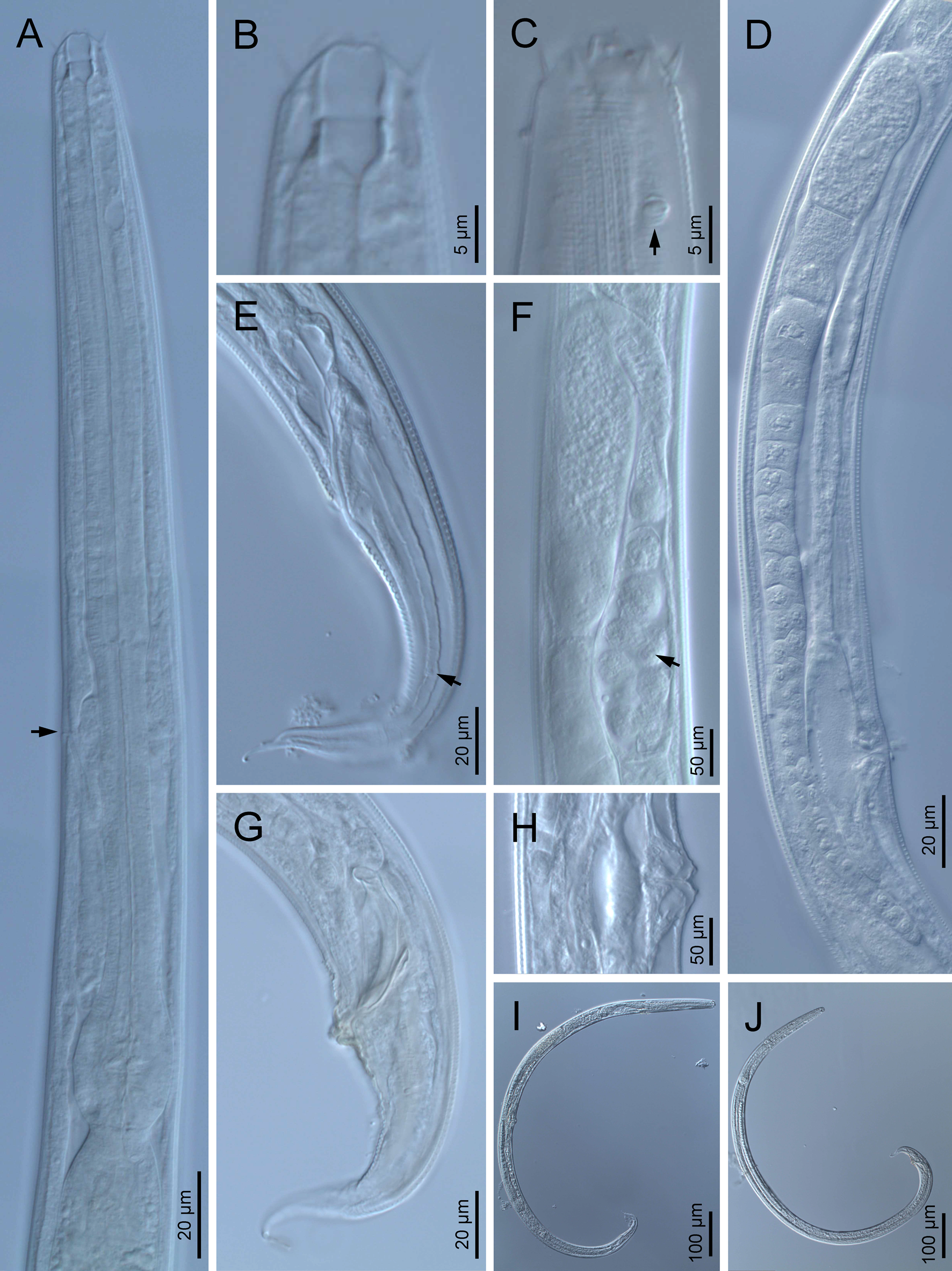
Fig. 1. Diastolaimus grossus (Truskova & Eroshenko, 1977) Andrássy, 1984 (LM): (a) Neck (arrow pointing the excretory pore); (b, c) anterior end at stoma and amphid (arrow) levels, respectively; (d) female reproductive system; (e) female posterior end (arrow pointing the phasmid); (f) spermatheca (arrow pointing the sperm); (g) male posterior end; (h) vagina; (i) entire female; (j) entire male.
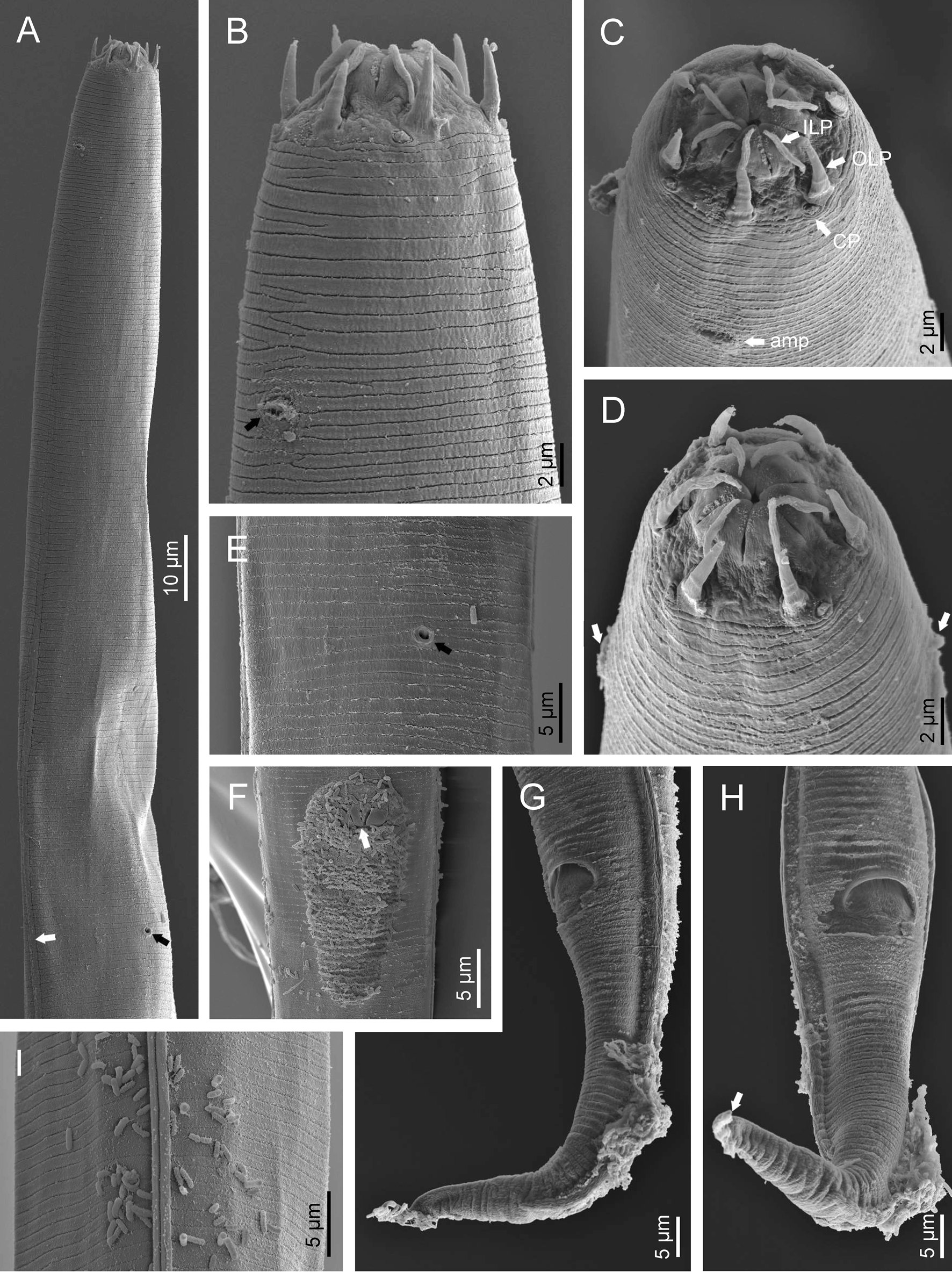
Fig. 2. Diastolaimus grossus (Truskova & Eroshenko, 1977) Andrássy, 1984 (SEM, female): (a) anterior end (black arrow pointing the excretory pore, white arrow pointing the deirid); (b–d) lip region in subventral, fronto-lateral and fronto-ventral views, respectively (arrows pointing the amphids); (e) excretory pore (arrow); (f) vulva (arrow); (g, h) posterior end (arrows pointing forked tail tip); (i) lateral field.

Fig. 3. Diastolaimus grossus (Truskova & Eroshenko, 1977) Andrássy, 1984 (SEM, male): (a, b) posterior end in ventro-lateral view (arrows pointing genital papillae); (c) tail end in lateral view (arrow pointing last genital papillae). ph, phasmid.
Female. Reproductive system didelphic-amphidelphic. Both ovaries reflexed near to oviduct junction, very long, surpassing the level of the vulva; anterior ovary is always longer than posterior one (1.1–1.2 times longer) and can be reflexed two times; oocytes first in two rows, primarily in short ovary reflection at vagina, then in only one row. Oviduct narrow and short with distinct narrowing spermatheca containing big spherical sperms. Uteri located on right-hand side of intestine; tubular, with thick walls and two times the corresponding body diameter long. Uterine eggs absent in the specimens examined. Vagina straight, occupying one-third (c. 31–40%) of body diameter. Vulva a transverse slit, with distinctly protruded lips that forms a vulval cone; advulval cuticle lacking differentiations. Rectum 1.6–1.7 times longer than anal body diameter, its anterior half swollen forming strong sphincter; anal lips not prominent. Tail conical, slightly ventrad curved, tapering very gradually and ending in a filiform forked terminus 8–9 μm long. Phasmids are located at 43–44% of tail length from anus.
Male. Reproductive system monorchic, with testis reflexed ventrad, on right-hand side of intestine. Spermatocytes first in two rows, then in only one row. Spicules paired and symmetrical, 5.7–6.0 times longer than wide, curved ventrad with rounded manubrium, cylindrical calamus, ventrad curved lamina with reduced dorsal hump and a well-developed ventral velum. Gubernaculum long, with thin manubrium and corpus with low central cuneus and well-developed triangular lateral crura. Fourteen pairs of genital papillae are present, eight subventral precloacal pairs and six postcloacal pairs arranged as follows: two subventral pairs located posterior to the level of cloacal aperture, one lateral pair located at lateral field, one subventral pair, one subdorsal pair at the posterior half of tail next to the phasmid and one lateral at the end of tail (six annuli before beginning of filiform part of tail end). Tail conoid and tapering gradually and curved ventrad, ending in a short terminally forked filiform part. Phasmids located at anterior half of tail, at 48% from anus.
Gower General Similarity coefficient of morphological characters. The analyses were based on morphometric and morphological characters including all available data of the redescribed populations in this article and taxa of the family Chambersiellidae previously published (see figs 4 and 5). Analyses of taxa of the family Chambersiellidae showed the identical formation of two main clusters, both with two subclusters for males and females. Main clusters clearly separate the genus Macrolaimus from the rest of the genera for Chambersiellidae. The second cluster involves the genera Chambersiella, Cornilaimus, Diastolaimus and Geraldius. Within subcluster A of cluster II, all species of the Diastolaimus genus are grouped, except D. papillatus in females and D. papillatus and D. mexicanus in males. Diastolaimus mexicanus is grouped together with D. grossus (Russian population) in females in subcluster A of cluster I. Cornillaimus furcillus is grouped closely with D. damalis in cluster I, subcluster II both for males and females. Males of D. grossus of Czech and Russian populations are grouped together in males and separated from other members of subcluster A of cluster I. Females of different populations of D. grossus are all grouped in cluster I, subcluster A. The Czech population of D. grossus is grouped with Bosnian and Russian populations with D. mexicanus.
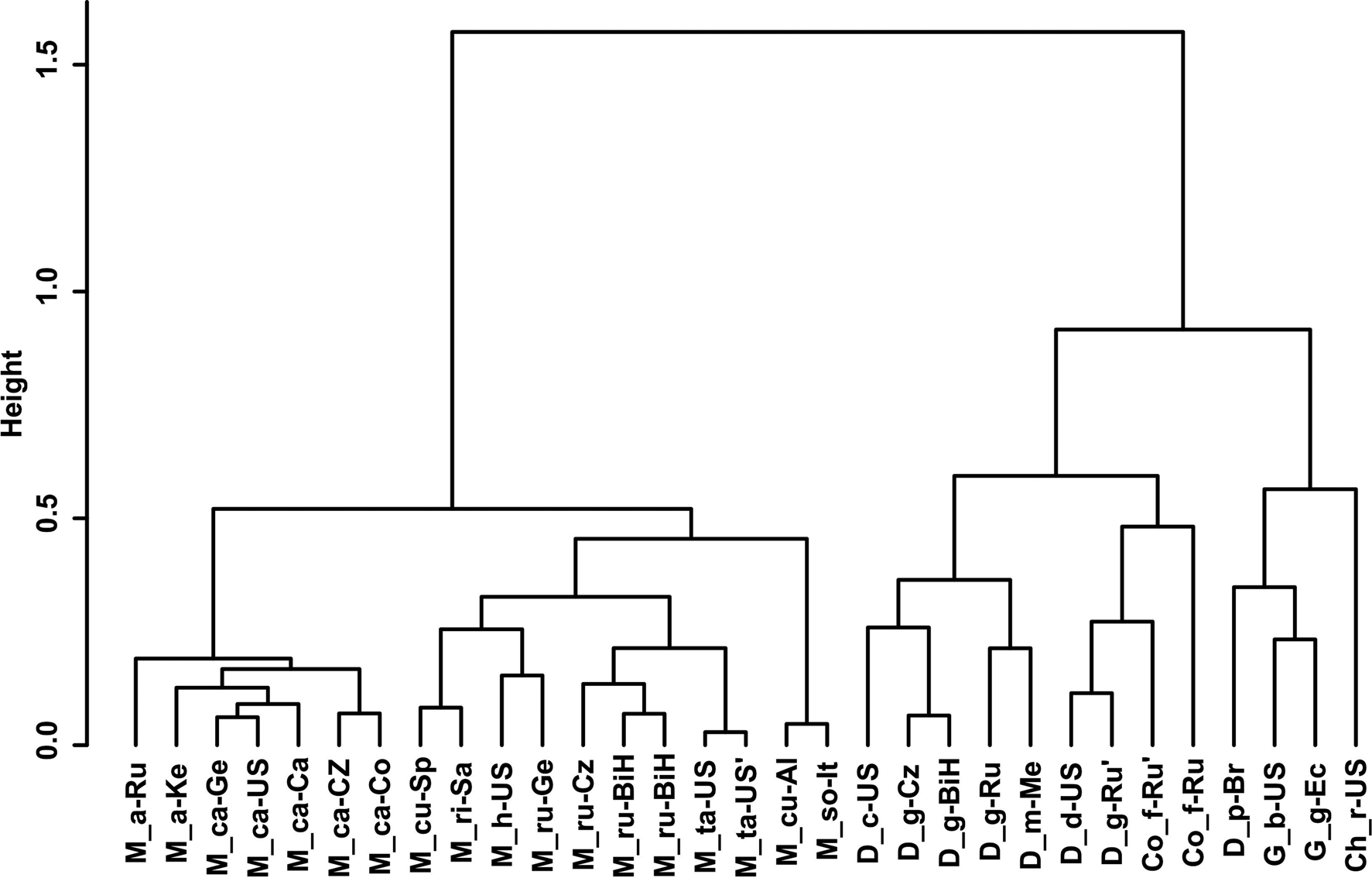
Fig. 4. Dendrogram from hierarchical clustering based on morphological characteristics of published members of the family Chambersiellidae (females).

Fig. 5. Dendrogram from hierarchical clustering based on morphological characteristics of published members of the family Chambersiellidae (males).
Other material examined
The material studied from Bosnia and Herzegovina (two females) is similar to the specimens from the Czech Republic, only showing slight differences in some morphometrical characters caused by their incomplete maturity based on not fully developed and functional vulva (see table 1).
This is the first record of the genus and species for the Czech Republic and for Bosnia and Herzegovina.
Molecular characterization
After sequencing the D2–D3 region of the 28S rRNA and 18S rRNA genes of D. grossus PCR product, partial sequences of 745 bp (OM691516) and 1684 bp (OM691517) were obtained, respectively.
Sequence similarity of D. grossus (OM691516) compared to Geraldius sp. (GU062821) was 85.3% (745 bp). However, the sequence similarity of D. grossus (OM691516) and Geraldius sp. (GU062821) with member of the subfamily Macrolaimiane Sanwal, 1971 M. canadensis (OM691512) was 66.4% (727 bp) and 67.5% (719 bp), respectively. There are no available sequences of any species of the genus Chambersiella. The BLAST homology search showed 100% similarity of the 18S rRNA gene sequence with F. grossa (KC242218), and close molecular similarity with the sequences of Acrobeles species A. complexus 88.82% (KU180671) and A. maximus 88.89% (EU306344). The similarity of D. grossus (OM691517) with members of the subfamily Macrolaiminae Sanwal, 1971 was as follows: M. canadensis (OM691513), 72.2% (1709 bp); M. ruehmi (OM691506), 78.0% (1682 bp); and M. crucis (OM691510), 75.3% (827 bp).
Two obtained 28S sequences of M. ruehmi (672 bp and 602 bp, respectively) were compared. The sequence similarity was 98.3%. Sequences of M. ruehmi (OM691514 and OM691515) were further compared to the M. arboreus sequence (MF996702) with 81.6% (614 bp) and 81.6% (599 bp) similarity, respectively, and to the M. crucis sequence (OM691509) with 72.2 (455 bp) and 72.8% (453 bp) similarity, respectively. Sequence similarity of M. ruehmi (OM691514) and M. canadensis (OM691512) was found to be 84.3%. A similar picture was revealed in comparison of the obtained 18S sequences (1682 bp) of M. ruehmi (OM691506) with the M. canadensis sequence (OM691513), at 94.3% (1709 bp), and with the M. crucis sequence (OM691510), which was also 94.3% (827 bp).
Remarks
Diastolaimus grossus differs from the original description of the holotype in some morphometrical characters (longer tail expressed by the ratio of tail length to body length (c) 12.6–14.7 vs. 19, vulva is located more anteriorly 51–54 vs. 59). From the male allotype in longer tail (c = 12.8–16.7 vs. 15–20 and longer spicule 33–39 vs. 30 μm). The main difference is in the number of mail tail papillae. Only six postcloacal papillae are present on the original description and possibly on precloacal vs. eight precloacal and six postcloacal pairs of papillae. The number of pairs of postcloacal papillae is coincident; however, their arrangement is different.
Discussion
About the genus Diastolaimus Rahm, 1928
The genus Diastolaimus was proposed by Rahm (Reference Rahm1928) for a new species, D. papillatus Rahm, 1928, with reference to the monotypic genus Chambersiella Cobb, 1920. Later, Andrássy (Reference Andrássy and Franz1984) synonymized the genus Diastolaimus Rahm, 1928 with the genera Fescia Truskova & Eroshenko, 1977 and Santafea Massey, 1963, all of which had identical morphological characters.
The genus contains five valid species. Diastolaimus mexicanus, described by Cid del Prado (Reference Cid del Prado2012) from Mexico, has morphology and morphometry identical to D. grossus. Thus, D. mexicanus is proposed as a new junior synonym of D. grossus.
List of species
-
Diastolaimus aculeatus (Daday, 1905) Andrássy, 1984
-
Syn. Cephalobus aculeatus Daday, 1905
-
Syn. Macrolaimus aculeatus (Daday, 1905) Thorne, 1937
-
Diastolaimus croca (Massey, 1963) Andrássy, 1984
-
Syn. Santafea croca Massey, 1963
-
Diastolaimus damalis (Massey, 1966) Andrássy, 1984
-
Syn. Santafea damalis Massey, 1966
-
Diastolaimus grossus (Truskova & Eroshenko, 1977) Andrássy, 1984
-
Syn. Fescia grossa Truskova & Eroshenko, 1977
-
Syn. D. mexicanus Cid del Prado, 2012 n. syn.
-
Diastolaimus papillatus Rahm, 1928
-
Syn. Chambersiella papilata (Rahm, 1928) Sanwal, 1960
Key to species identification
1a Body more than 1.9 mm………2
1b Body less than 1.9 mm………3
2a Spicules with not swollen manubrium; spicules c. 2.5 times the gubernaculum length………D. aculeatus
2b Spicules with swollen manubrium; spicules twice the gubernaculum length………D. papillatus
3a Cephalic papillae longer than the anterior part of gymnostom; stoma narrow………D. damalis
3b Cephalic papillae shorter than the anterior part of the gymnostom; stoma wide………4
4a Posterior part of gymnostom very short; spicules 52 μm long with manubrium ventrad bent………D. croca
4b Posterior part of gymnostom slightly shorter than the anterior part; spicules 30–40 μm long with manubrium not ventrad bent……… D. grossus
Phylogenetic position of the genus Diastolaimus and its relatives
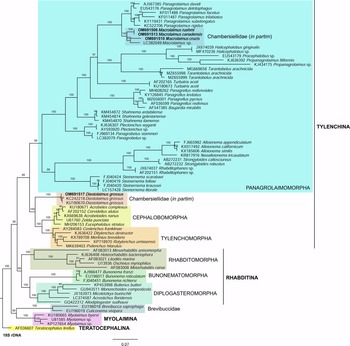
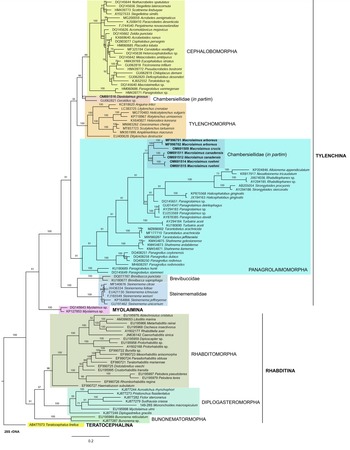
In the phylogenetic study of Nadler et al. (Reference Nadler, De Ley and Mundo-Ocampo2006), Fescia (junior synonym of Diastolaimus) and Macrolaimus were always monophyletic with strong support (100% by maximum parsimony (MP) bootstrap) and in the MP consensus tree were more closely related to members of the clade including Cephalobomorpha and Tylenchomorpha. However, their position among sampled nematodes varied according to tree inference method. De Ley & Blaxter (Reference De Ley, Blaxter, Cook and Hunt2004) considered the structure of sensory organs of the members of the family Chambersiellidae as plesiomorphic compared to Cephalobomorpha or Panagrolaimomorpha. Holovachov et al. (Reference Holovachov, Camp and Nadler2015) considered that their large 18S + 28S dataset appears to contain insufficient phylogenetic signal for unambiguously resolving the relationship of F. grossa. In five out of eight analyses, it was sister taxon to Cephalobomorpha + Tylenchomorpha, but with varying support values. However, for the purposes of the analyses of the unpartitioned secondary structure alignment, Fescia was a part of the Panagrolaimomorpha clade (Holovachov et al., Reference Holovachov, Camp and Nadler2015).
Phylogenetic analysis based on 18S rDNA (fig. 6) and 28S rDNA (fig. 7) fragments revealed a clear polyphyletism of the family Chambersiellidae as well as the subfamily Macrolaiminae Sanwal, 1971, which positively corresponds with the results of Grow–Ward analysis. All the Macrolaimus species, which represent a more evolutioned species with a less complicated structure of sensory organs (Abolafia et al., Reference Abolafia, Ruiz-Cuenca, Foit and Čermák2018; fig. 8), appear as monophyletic taxon in the infraorder Panagrolaimomorpha. On the other hand, the genus Diastolaimus (Macrolaiminae cf. Andrássy, Reference Andrássy and Franz1984, Reference Andrássy, Csuzdi and Mahunka2005) appears located together with the genus Geraldius Sanwal, 1971 (Charbersiellinae). According to this, the genus Diastolaimus is transferred to the subfamily Charbersiellinae, which represent taxa including ancestral plesiomorphic characters appearing as a sister taxon of Cephalomorpha, while the subfamily Macrolaiminae, with the only genus Macrolaimus, is located in the family Panagrolaimidae.
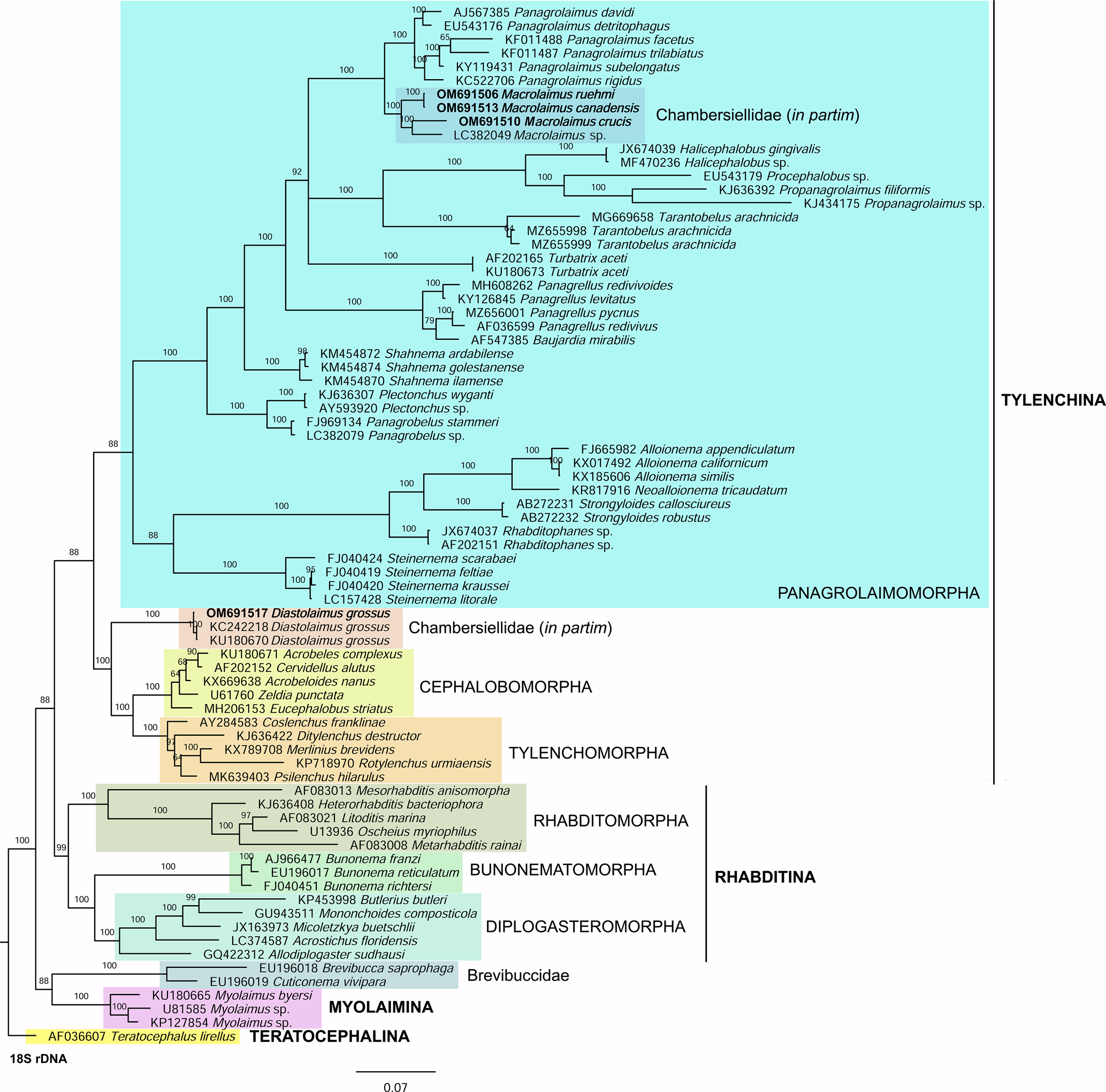
Fig. 6. The Bayesian tree inferred from rhabditid species based on sequences of the 18S rDNA region. Bayesian posterior probabilities (%) are given for each clade. Scale bar shows the number of substitutions per site.

Fig. 7. The Bayesian tree inferred from rhabditid species based on sequences of the 28S rDNA region. Bayesian posterior probabilities (%) are given for each clade. Scale bar shows the number of substitutions per site.
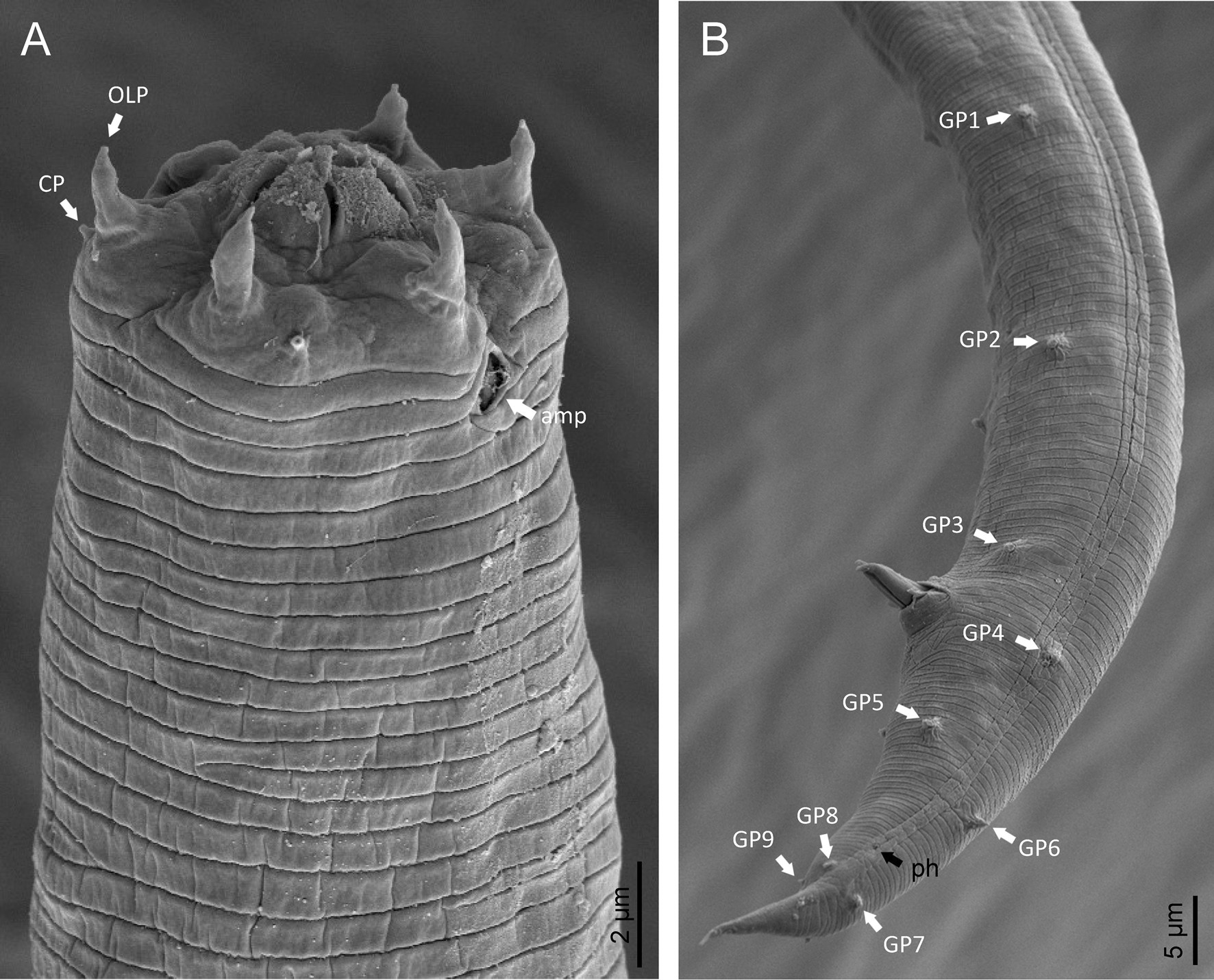
Fig. 8. Lip region (a) and male posterior end (b) of Macrolaimus crucis Maupas, 1900 from coastal sand dune in Alicante (Spain).
Abolafia et al. (Reference Abolafia, Ruiz-Cuenca, Foit and Čermák2018) and Nadler et al. (Reference Nadler, De Ley and Mundo-Ocampo2006) connected the number of precloacal papillae and number of reproductive branches with the phylogenetic position. Species with a didelphic female reproductive system represent an ancestral (plesiomorphic) character, whereas species with a monodelphic reproductive system represent a derived (apomorphic) character (Chitwood & Chitwood, Reference Chitwood and Chitwood1950; Lorenzen, Reference Lorenzen1978, Reference Lorenzen1981). In the 28S phylogenetic tree (fig. 7), D. grossus (X645-28S) is placed in one clade with Geraldius sp. (GU062821). Both genera share ancestral characters including high number of papillae and didelphic reproductive system in females. The only species of the genus Geraldius Sanwal, 1971, G. bakeri, bears seven pairs and eight pairs of postcloacal papillae. Other related species C. rodens also has a high number of precloacal papillae – six pairs and eight pairs of postcloacal, despite monodelphic female reproductive system. Diastolaimus croca has 13 pairs of caudal papillae, four pairs subventral, one pair lateral preanal, one pair postanal and lateral, four pairs postanal subventral and three pairs postanal and subdorsal. Based on the Gower–Ward method (figs 4 and 5), the key grouping morphological character besides number of caudal papillae is also the level of complexity of labial structures. All the genera with higher level of complexity of labial structures are grouped together, regardless of the number of reproductive branches in females. We can assume that the different/lower number of precloacal papillae in the original description of D. grossus provided by Truskova & Eroshenko (Reference Truskova and Eroshenko1977) is not the difference between species, but just that the precloacal papillae was not originally observed.
The latest described species D. mexicanus (Cid del Prado, Reference Cid del Prado2012) is very similar to D. grossus, especially with respect to females (see fig. 3). Males are slightly different in some morphometrical characters: number of pairs of genital papillae (13 in D. mexicanus vs. 14 in D. grossus), more posteriorly located excretory pore (158 vs. 128 μm; however, an illustration in Cid del Prado (Reference Cid del Prado2012) shows that its position is similar), phasmid–cloaca distance (18 vs. 41 μm). However, with respect to the number of genital papillae, SEM illustrations in the original description are not clear and perhaps some of the genital papillae were not observed in D. mexicanus. The excretory pore is in the same location in both species according to the illustration of D. mexicanus. With respect to the phasmid–cloaca distance, at least some measurements of the male phasmid position are incorrect in D. mexicanus since the description states 18 μm while the drawing gives the measurement as 28 μm (= 40%) – this range is too large and perhaps the phasmids were confused with genital papillae in some measurements. According to this, we proposed D. mexicanus as a junior synonym of D. grossus.
Comments on the distribution and ecology of D. grossus and its relatives
To date, D. grossus is known only from four countries: Russia (Truskova & Eroshenko, Reference Truskova and Eroshenko1977), Ukraine (Nadler et al., Reference Nadler, De Ley and Mundo-Ocampo2006), the Czech Republic and Bosnia and Herzegovina (present study). All the members of the subfamily Chambersiellinae can be considered as epiphytic nematodes living on the bark of various tree species and can survive rapid and long dehydration (Cobb, Reference Cobb1920; Rahm, Reference Rahm1928). Diastolaimus grossus was originally extracted from the bark of fir tree twigs. Bosnian specimens were also extracted from the bark of a coniferous tree (P. nigra); only the Czech population was found on the bark of a broadleaf tree (Salix sp.). With respect to other members of the genus Diastolaimus, almost all of the species are associated with tree surface and are mainly distributed in the western hemisphere: D. croca and D. damalis in association with white fir, juniper and pine tree in North America (Massey, Reference Massey1963, Reference Massey1966); D. papillatus in South America (Brazil) in moss on orange leaves (Rahm, Reference Rahm1928). Daday (Reference Daday1905) did not note any details about sampling site and associations for D. aculeatus except that the location lake was close to Estia Postillon. Chambersiella rodens was found in many parts of the eastern US in association with various trees (Cobb, Reference Cobb1920) and Geraldius backeri from the bark of the oak tree in Ontario, Canada (Sanwal, Reference Sanwal1957). Except for D. grossus, the second European member of the subfamily Chambersiellinae is Cornilaimus furcillus, which was found in Far East Russia in association with the plant Cornus (Chamaepericlymenum) canadensis L.
Members of the subfamily Macrolaiminae are living both in soil and on the bark of various trees as epiphytic organisms. Macrolaimus crucis was found three times in different kinds of soil (Abolafia & Peña-Santiago, Reference Abolafia and Peña-Santiago2014); M. richteri was extracted from dry sandy soil in rocky desert; M. arboreus was found originally on the bark of fir in Russia, and secondary in soil in the rhizosphere of a pine tree in Iran. Based on all known findings of species, M. ruehmi generally prefers general broadleaf trees, but it was, however, found repeatedly in one location to be associated with Pinus sylvestris (Abolafia et al., Reference Abolafia, Ruiz-Cuenca, Foit and Čermák2018). Macrolaimus canadensis has not been found to be associated with any broadleaf tree yet (Fuchs, Reference Fuchs1938; Massey, Reference Massey1974; Abolafia et al., Reference Abolafia, Ruiz-Cuenca, Foit and Čermák2018). Macrolaimus canadensis and M. ruehmi seem to be obligatory epiphytic organisms associated with trees.
Members of the subfamily Chambersiellinae can be considered to be less evolved in comparison with Macrolaiminae, especialy their way of life and ecological niche (epiphytic/tree association) can be perceived as an ancestral bionomical character. Soil association probably came later, and can be facultative to a certain degree. The occurrence of the same nematode species in a different niche is well known and spread among different taxonomic groups of nematodes, such as: Pristionchus (Sudhaus & Fürst von Lieven, Reference Sudhaus and Fürst von Lieven2003), Halicephalobus (Taulescu et al., Reference Taulescu, Ionică, Lazar, Pavaloiu, Cora, Amorim, Catoi and Roccabianca2016), Bursaphelenchus (Čermák et al., Reference Čermák, Vieira, Cudejkova, Gaar, Tomankova, Mikuskova, Eisenback and Mota2014), Prionchulus (Andrássy, Reference Andrássy2009). This suggests the facultative transition between bark and soil environment can be beneficial for the species survival and improve its competitive strategies.
Acknowledgements
SEM pictures were obtained with the assistance of technical staff and equipment of ‘Centro de Instrumentación Científico-Técnica (CICT)’ from the University of Jaén. Molecular data were acquired with the help and support of Kateřina Tománková from the Laboratory of Molecular Biology, Central Institute for Supervising and Testing in Agriculture. We are grateful to Ladislav Háněl from the Institute of Soil Biology, Czech Academy of Science, for valuable comments and taxonomical advice.
Financial support
The authors thank the University of Jaén, Spain, for financial support received for the Research Support Plans ‘PAIUJA 2019/2020: EI_RNM02_2019’ and ‘PAIUJA 2020/2021: EI_RNM02_2020’; the European Regional Development Fund for financial support received via project ‘FIT’ (Pharmacology, Immunotherapy, nanoToxicology) (CZ.02.1.01/0.0/0.0/15_003/0000495); and the Czech Development Agency (CZDA) for their support via the project ‘Food Safety Improvement in Bosnia and Herzegovina’.
Conflicts of interest
None.
Ethical standards
All procedures contributing to this study comply with the ethical standards of the relevant national and institutional guides on the care and use of animals.