What you need to know
Background: Coeliac disease (CD) is exacerbated by gluten proteins in wheat and related cereals that have been characterised.
Findings: CD-toxic peptides contain glutamine and proline residues which if singly substituted in CD-toxic peptides decreases or if jointly changed abrogates CD toxicity; certain naturally occurring wheat glutens do not contain CD-toxic motifs.
Implications for patient care: The findings permit the potential generation of (a) a novel cereal that produces flour with the baking and sensorial characteristics of wheat that we hypothesise is non-toxic to subjects with CD and (b) non-GM wheat, using standard breeding techniques that may be CD safe.
Coeliac disease (CD) is a common T cell inflammatory disorder that affects 1–2 % of individuals worldwide. The management of the condition involves a gluten-free diet (GFD) with avoidance of wheat and related cereals(Reference Ciclitira1). A GFD is restrictive, resulting in reduced quality of life(Reference Deepak, Berry and Kochlor3,Reference Ciclitira, Evans and Fagg4) . The condition is exacerbated by gluten proteins from wheat and homologous proteins from barley, rye and potentially oats. CD manifests in genetically predisposed individuals who have the genes that encode the HLA variants DQ2 or DQ8. Adaptive immune response in these individuals is activated by the interaction between gluten peptides bound on HLA-DQ2/HLA-DQ8 molecules on antigen presenting cells and T cell receptors on the surface of small-intestinal CD4+ T cells. Fully activated inflammatory response mediated by T cells leads to tissue damage in the small intestine as an atrophy of the intestinal villi and crypt proliferation. There is evidence that an additional pathway is involved in CD pathogenesis supporting an innate enterocyte as well as an adaptive T cell response. Initial epithelial damage was observed 1 h following ingestion of gluten, which is too early for a fully activated T cell response(Reference Ciclitira, Evans and Fagg4). Moreover, it has been shown that some gluten peptides cause tissue damage without stimulating CD4+ T cells. It is established that the adaptive immune response is through CD4+ T cells and an innate immune response mediated by IL-15 co-operates to cause small-intestinal tissue damage(Reference Korneychuk, Ramiro-Puig and Ettersperger5).
Gluten is principally composed of gliadins, low and high molecular weight glutenins, all three classes of which have been reported to be CD toxic(Reference Ciclitira, Evans and Fagg4–Reference Molberg, Solheim Flaete and Jensen8). Previous studies suggested that gliadins tend to be more toxic than glutenins and, therefore, have been the subject of many CD toxicity research studies(Reference Korneychuk, Ramiro-Puig and Ettersperger5). Gliadins can be further divided into α, γ and ω fractions, according to their electrophoretic mobility. Studies have suggested that within gliadins, α-gliadins are the most CD toxic(Reference Dewar, Amato and Ellis7). Proline (P) and glutamine (G) amino acid (AA) residues are of particular relevance for CD pathogenesis. The α-gliadins have high contents of both AA, especially in their N-terminal part(Reference Sollid, Qiao and Anderson9). We considered the effect of digestion that is known to result in the generation of a 33mer long fragment from the N-terminal part of α-gliadins, which related to our choice of peptides for evaluation. Such a fragment includes several CD-toxic epitopes (DQ2.5-glia-α2, DQ2.5-glia-α1 and DQ2-glia-α1b), which bind well with HLA-DQ2 and HLA-DQ8 molecules and are recognised by gluten-sensitive T cells(Reference Molberg, Solheim Flaete and Jensen8–Reference Ellis, Pollock and Engel10).
As a consequence of nucleotide substitutions of α-gliadin genes during the evolution of cereals, naturally existing AA sequence variations of α-gliadins have been found within wheat species. Such large-scale analysis of full-length α-gliadin complementary DNA (cDNA) became possible because the new sequencing platform enabled each cDNA to be sequenced in the respective entire length(Reference Zhang, Ciclitira and Messing11). This aspect is critical because α-gliadins have a repetitive sequence motif preventing the reconstruction of individual cDNA from overlapping sequence information. Taking such an approach, one can ask whether natural variations among wheat species can be used to identify α-gliadin genes that encode proteins with reduced content of T cell epitopes without loss of bread-making qualities. Then, the question remains whether these variants of T cell stimulatory epitopes have reduced immunogenicity for CD-affected subjects. Given a list of known toxic epitopes to date(Reference Sollid, Qiao and Anderson9), previous studies have shown that a single AA substitution within a T cell epitope may be able to eliminate the immunogenicity of gluten peptides and it was reported these CD-toxic epitopes are not broken down by inter-luminal digestive enzymes(Reference Mitea, Salentijn and van Veelen12–Reference Molberg, Uhlen and Jensen14). Therefore, we investigated the immunogenic potential of T cell epitopes and their naturally existing variants with T cell proliferation assays with CD small-intestinal gluten-sensitive T cells. Knowing the heterogenic response of cell lines from different patients on the same epitopes, our study was carried out using polyclonal gluten-specific T cell lines isolated from duodenal biopsies. An in vitro model involving small-intestinal organ culture was used as an additional method to examine CD toxicity of each α-gliadin peptide that enabled us to mimic the environment in the human intestine more closely, following generation of peptides, due to digestion by luminal enzymes, including pepsin and trypsin.
In our study, we sought to characterise CD-toxic gluten epitopes and their naturally existing sequence variants. Screening for sequences within gluten proteins that are non-toxic would have significant implications in utilising naturally occurring variants, including the potential to design CD non-toxic wheat proteins, with reduction or absence of CD toxicity. Furthermore, genes encoding specifically selected CD non-toxic sequences of gluten proteins could be expressed in a gluten-free cereal. This would potentially enable production of flour with the sensorial and baking characteristics of wheat but with a preserved CD non-toxic profile, although the rheological characteristics of the resultant flour would need to be assessed.
Methods
Synthetic peptides
Peptides including variations of known epitopes were synthesised as 16mers with 95 % purity by Selleck Chemicals (www.selleckchem.com). The choice of the peptides from α-gliadin followed previously reported CD-toxic epitopes(Reference Molberg, Solheim Flaete and Jensen8–Reference Ellis, Pollock and Engel10,Reference Mitea, Salentijn and van Veelen12–Reference Molberg, Uhlen and Jensen14,Reference Molberg, McAdam and Lundin18) .
Patients
Small-intestinal biopsies were taken during follow-up evaluation in subjects with suspected or known CD. Out of fifteen, six were on a normal diet and nine on a GFD (Table 1). In line with the British Society of Gastroenterology Guidelines for the management of Coeliac Disease(Reference Ludvigsson, Bai and Biagi15), subjects were diagnosed according to these guidelines. All the subjects (n 15) read and signed the informed consent form. The study was approved by the St Thomas’ Hospital Research Ethics Committee institutional review board, reference numbers 05/Q0207/167. Participating patients were typified as HLA-DQ2 positive and in one case as DQ8 positive. The subjects included eleven females and four males (Table 1) with a median age of 41 years and an age range from 27 to 64 years. Biopsies were taken from the second part of the duodenum, a common procedure required for the management of the condition in adults(Reference Ciclitira1).
Table 1. Details of coeliac disease volunteers from whom small-intestinal biopsies were taken for research purposes*
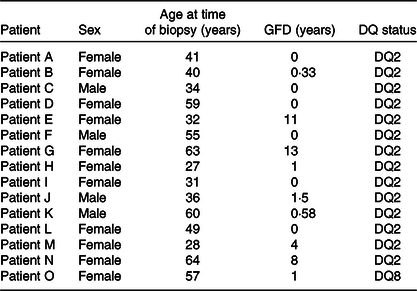
GFD, gluten-free diet.
* Their sex, age, DQ status and period on a GFD were recorded on the day of the procedure.
Preparation of antigens
Industrial gluten (Vital Wheat Gluten, Roquette UK Ltd) and ovalbumin were peptic tryptic (PT) digested following Frazer’s method(Reference Frazer, Fletcher and Ross16) to serve, respectively, as positive and negative controls. PT digests were further deamidated using a deamidation mix as follows: 100 μg/ml PT-digested gluten or ovalbumin and 2 mm calcium chloride. The mixtures were incubated at 37°C for 4 h. PT and tissue transglutaminase pre-treated gluten were used for restimulation in T cell studies and as positive control in proliferation assays and organ culture. Ovalbumin was used as a negative control in organ culture assays. In all the proliferation assays employing ovalbumin, this control protein always resulted in a negative stimulation index (SI < 2). Therefore, medium only subsequently served as a negative control.
Establishment of small-intestinal T cell lines
The methods were undertaken as previously described(Reference Dewar, Amato and Ellis7,Reference Molberg, Uhlen and Jensen14) . Small-intestinal biopsies from CD-affected subjects were challenged overnight in an organ culture chamber in contact with 5 mg/ml PT-digested gluten solution. Lymphocytes were isolated by mechanical disruption of biopsies the next day and passed through a 70 μm cell filter (Falcon; Becton Dickson Ltd). T cells were cultured in the Roswell Park Memorial Institute (RPMI) 1640 tissue culture medium supplemented with 10 % heat inactivated autologous serum, plasmocin 2·5 μg/ml; (Invivigen Ltd, Cayla), amphotericin B (0·25 mg/ml; PAA Cell Culture Company) and Hepes buffer. The medium was refreshed every 2–3 d, and IL-2 (17903, Sigma) was added as a growth factor. Every seventh day, cells were restimulated with autologous irradiated peripheral blood mononuclear cells (22 Grey) that had been previously incubated with PT-digested and tissue transglutaminase-treated gluten.
T cell proliferation assays
The T cells used were obtained following 1–4 weeks in culture prior to testing in T cell proliferation assays, depending on the growth of the cells. Complex antigen (gluten) was incubated with antigen presenting cells (5 × 106); plates were incubated for 48 h. Tritiated thymidine was added (NET355, Perkin Elmer) for 18 h prior to harvesting with a Tomtec Cell Harvester. The level of thymidine incorporation in the T cell DNA was measured using a Wallac 1450 Microbeta Plus liquid scintillation counter. SI were calculated following the equation:
 $${\rm{SI}} = {\rm{mean\; counts\; per\; min\; for\; T\; cells}} + {\rm{antigen}}/{\rm{mean\; counts\; per\; min\; for\; T\; cells}} + {\rm{APC}}.$$
$${\rm{SI}} = {\rm{mean\; counts\; per\; min\; for\; T\; cells}} + {\rm{antigen}}/{\rm{mean\; counts\; per\; min\; for\; T\; cells}} + {\rm{APC}}.$$An SI of 2 or more was considered positive.
Duodenal biopsy organ culture
Paired duodenal biopsies from CD patients were placed in separate organ culture dishes containing either 1 mg/ml of PT-digested gluten as a positive control(Reference Dewar, Amato and Ellis7,Reference Zevallos, Ellis and Ciclitira17) or 200 μg/ml of individual peptide (peptides 3, 5, 6 and 9). Following overnight incubation, the biopsies were fixed in 10 % formaldehyde solution and after at least 48 h embedded in paraffin (Thermo Shandon Histocentre 3 Embedding Station). Tissue blocks were cut to a 5 μm thickness and stained with haematoxylin–eosin. Sections were microscopically examined, selecting at least three villi in order to measure the enterocyte surface-cell height (ESCH), one third of the way down the villi, ten continuous enterocytes were measured on each villus, giving a total of thirty ESCH (haematoxylin–eosin) measurements for each section. Where villi were not able to be identified but the enterocyte layer was clear, measurements were taken from the middle of the section. Means of ESCH for the sample groups were compared. An ANOVA statistical analysis was used to compare mean ESCH between sample groups.
Results
Selection of sequence motifs for epitope analysis
A common path to identify epitopes has been the use of peptide arrays(Reference Szymczak, Kuo and Mrksich19). Although one can argue that in such cases peptides do not represent natural epitopes because their sequences might be hidden due to the three-dimensional structure of proteins and not immunogenic in the native protein, this does not apply to gluten proteins because they are digested and hydrolysed into many small peptide fragments, when they reach the small intestine. Therefore, one can use synthetic peptides to simulate gluten epitopes that affect CD patients. Previously, α-gliadins from ten diverse wheat cultivars had been sequenced and assembled using the PacBio system(Reference Zhang, Ciclitira and Messing11). These assembled sequences were aligned to identify variations in sequence motifs (Fig. 1). Peptides including variations of known epitopes were synthesised as 16mers with 95 % purity as described under Methods. In this study, we present the results of toxicity testing of α-gliadin peptides marked as peptides 1, 3, 5, 6 and 9 for their simplicity of labelling in the text and tables. Peptide 1 harbours CD-toxic gluten epitopes (DQ2.5-glia-α-1a or DQ2.5-glia-α2); peptide 3 is the deamidated counterpart at positions p65 and p72; peptides 5 and 6 are deamidated naturally existing variants of peptide 1; peptide 9 does not naturally exist, but was designed to have glutamine at p63, p65 and p70 mutated to lysine (Table 2). Peptides 1, 3, 5, 6 and 9 were tested for recognition by eleven polyclonal T cell lines, specific for gluten (Table 2). Proliferation assays for each antigen were performed in triplicate. CV of triplicates were calculated, and CV 0·2 or less was considered as acceptable variability. Results are presented as SI with corresponding arithmetic mean and one standard deviation (Table 3). The results for Patient O, who was HLA-DQ8 in Table 4, were no different from the others who had the HLA-DQ2 allele.

Fig. 1. Snapshot alignment of DQ2.5-glia-α1 and DQ2.5-glia-α2 region in ten wheat cultivars used in a previous study(Reference Zhang, Ciclitira and Messing11).
Table 2. Amino acid sequences for α-gliadin peptides and substituted variants used in the study
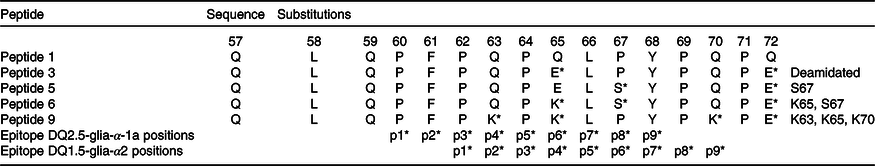
* Amino acids within the immunodominant p57–p71 peptide are present and the position of their binding into HLA-DQ2 groove (p1, p2…).
Table 3. Results of stimulation indices (SI) of eleven proliferation assays; eleven gluten-sensitive T cell lines were tested with medium only, peptic tryptic-digested gluten and peptides 1, 3, 5, 6 and 9
(Mean values and standard deviations)

* SI for individual antigens, with the corresponding arithmetic means and standard deviations. Note that SI greater than 2 were considered as positive.
Table 4. Comparison of mean enterocyte surface-cell height (ESCH) (μm) in coeliac disease patients; paired biopsies were left overnight in the organ culture system under seven conditions: medium only, ovalbumin, peptic tryptic-digested gluten, peptides 3, 5, 6 and 9*

* P values show if the difference between ESCH is statistically significant when compared with medium.
Deamidation of dominant gluten epitopes in α-gliadins
Peptide 1 (QLQPFPQPQLPYPQPQ) is an immunodominant peptide harbouring overlapping T cell epitopes; DQ2.5-glia-α1a, DQ2.5-glia-α2 and part of DQ2.5-glia-α1b (from a nomenclature of CD-toxic epitopes)(Reference Sollid, Qiao and Anderson9). To confirm the importance of deamidation at position p65 and p72 of α-gliadin, a deamidated counterpart (peptide 3) was tested in ten gluten-specific cell lines with proliferation assays determined. A table of responses for both peptides is shown in Table 3 with the SI.
Even though peptide 1 and its deaminated counterpart (peptide 3) were evaluated in previous studies as immunodominant peptides, they had not been tested positive (SI > 2) in all the tested T cell lines. Approximately half of gluten-specific cell lines (7/15) recognise peptide 1 or peptide 3 (Fig. 2).
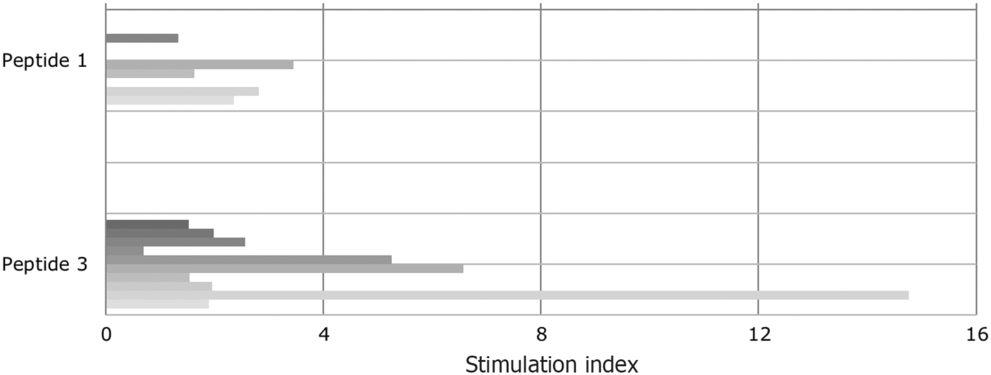
Fig. 2. Proliferative response of T cell lines to immunodominant peptide 1 (epitope DQ2.5-glia-α1a positions and DQ2.5-glia-α2) and to peptide 3, the deamidated counterpart. Tissue transglutaminase increased the stimulatory capacity. The patients are vertically sequentially presented: peptide 1: patients (i) I, (ii) E, (iii) D, (iv) B and (v) A; peptide 3: patients (i) K, (ii) J, (iii) C, (iv) H, (v) G, (vi) E, (vii) D, (viii) J, (ix) B and (x) A.
SI varied from 0·7 to 14·76. This confirms the different sensitivity of T cells obtained from CD-affected subjects to specific epitopes. Notably, indices increased when peptide 1 had been deaminated. In the case of four T cell lines (patients B, E and G), the SI for peptide 3 were high and well above the cut-off for positivity (SI > 2).
Amino acid substitutions within T cell epitope in α-gliadins
The results involving peptides 5, 6 and 9 with AA incorporating substitutions in the T cell epitope were compared with those involving the immunodominant peptide 3. We wondered if the T cell response with these substitutions would be abrogated (Fig. 3). To address the question, in each peptide, higher numbers of substitutions were introduced. In peptide 5 (QLQPFPQPELSYPQPE), proline (P) at position 67 of α-gliadin was substituted with serine (S). This peptide triggered a positive response of T cells in two out of six patients. This was in contrast with the deamidated immunodominant peptide (peptide 3-QLQPFPQPELPYPQPE), with which the immune response was down-regulated, indicating the importance of proline at p67 in triggering the coeliac reaction in some patients. However, this peptide failed to be completely non-immunogenic for all the coeliac patient’s gluten-sensitive T cells that were assessed.

Fig. 3. Proliferative response of T cell lines to different α-gliadin peptides: peptide 3, peptide 5, peptide 6 and peptide 9. With every further peptide, one amino acid substitution is introduced. With two amino acid substitutions, T cell response is completely abrogated. The patients are vertically sequentially presented. Peptide 3: patients (i) K, (ii) J, (iii) I, (iv) H, (v) G, (vi) E, (vii) D, (viii) C, (ix) B and (x) A; peptide 5: patients (i) I, (ii) F, (iii) E, (iv) E, (v) B and (vi) A; peptide 6: patients (i) K, (ii) J, (iii) I, (iv) H, (v) G (vi) E (vii) B and (viii) A; peptide 9: patients (i) I, (ii) G, (iii) C, (iv) B and (v) A.
Peptide 6 (QLQPFPQPKLSYPQPE) compared with peptide 5 has blocked p65 glutamine from deamidation: glutamic acid (E) at position 65 was substituted with lysine (K) and proline (P) at position 67 with serine (S). Importantly, the response of gluten-specific T cells was completely abrogated in all the tested cell lines – none of the eight cell lines yielded a positive result. The result points to the importance of p65 deamidated glutamic acid in triggering the coeliac T cell response.
Finally, peptide 9 (QLQPFPKPKLPYPKPQ) had mutated deamidation point glutamine at both p65 and p70 (Q–K) and Q–K mutation at p63. Peptide 9 was tested with five polyclonal CD gluten-sensitive T cell lines. The SI for this peptide was negative (SI < 2) in five out of five samples (100 %).
The same peptides tested that had been in T cell proliferation assays were assessed in small-intestinal organ culture as the next step to examine their CD toxicity. Mean ESCH were measured as a marker of the CD toxicity of proteins/peptides. Table 4 shows the combined ESCH for each patient (L, M, N and O) under experimental conditions. Measurements of mean ESCH following overnight incubation of small-intestinal biopsies with medium only/ovalbumin were compared with measurements of mean ESCH after incubation of biopsies with PT-digested gluten with different peptides for each patient (Fig. 4).

Fig. 4. Results of overnight incubation of paired biopsies with medium only, peptic tryptic (PT)-digested gluten (GLU), PT-digested ovalbumin (OVA) and peptides 3, 5, 6 and 9. Figures show median enterocyte surface-cell height (ESCH), first and third quartiles, including minimum and maximum values for each patient.
In all tested samples, mean ESCH for biopsies cultured with digested gluten were significantly reduced in comparison with medium or ovalbumin. In specimens from patient N, mean ESCH from biopsies cultured with ovalbumin was significantly lower than biopsies cultured with medium alone. However, mean ESCH in this sample for ovalbumin was still significantly higher compared with digested gluten (P = 0·001), peptide 3 (P < 0·0001) and peptide 5 (P < 0·0001). In all other samples, mean ESCH between medium alone and ovalbumin did not reach statistical significance.
In all biopsies cultured with peptide 3, a reduction in ESCH was noted, indicating peptide 3 indeed triggers coeliac reaction in organ culture. There was a trend towards reduced mean ESCH when incubated with peptide 5 in comparison with medium alone and ovalbumin. Nevertheless, when tested on biopsies from patient O, no significant reduction of ESCH was observed. There was no difference in the result from the subject who was HLA-DQ8 compared with HLA-DQ2. This is in accordance with T cells results that peptide 5 only reduces coeliac response in some but not all patients.
However, when CD tissue incubated with the organ culture medium only was compared with the tissue incubated with peptide 6, there was no statistically significant difference (P < 0·005). Likewise, there was no statistically significant difference observed when cultured with peptide 9 (P < 0·05) (Table 4). The organ culture results point to the conclusion that deamidated glutamic acid (E) is crucial in triggering coeliac immune response, consistent with previous studies(Reference Molberg, McAdam and Lundin18).
Discussion
CD is a lifelong condition that affects 1–2 % of subjects worldwide. The only generally accepted treatment is a strict lifelong GFD, with avoidance of wheat, rye and barley. This is restrictive with resultant decreased quality of life(Reference Nachman, Mauriño and Vazquez2,Reference Deepak, Berry and Kochlor3) . Compliance to the GFD is challenging in both adults and children(Reference Nachman, Mauriño and Vazquez2). All three classes of wheat gluten of wheat proteins, gliadins, low molecular weight glutenins and high molecular weight glutenins exacerbate CD. The pathogenesis involves apparent adaptive T cell and innate enterocyte gluten-induced damage to the small-intestinal mucosa.
There is a great expectation for alternative therapies among patients and their families(Reference Hall, Rubin and Charnock20–Reference Branchi, Tomba and Ferretti23) with the suggestion that wheat deficient in gliadin may provide a promising tool for the treatment of CD(Reference Frisoni, Corazza and Lafiandra24).
We assessed five variants of α-gliadin immunodominant CD-toxic peptides synthesised as 16mers in CD T cell stimulation assays involving polyclonal gluten-sensitive T cells generated from duodenal biopsies from CD-affected individuals. The results revealed the mutation of proline (P) to serine (S) of p67 in α-gliadin peptides reduced immunogenicity and further blocking glutamine at p65 by mutation to lysine completely abrogated immunogenicity, suggesting that such peptides would have reduced CD toxicity. We subsequently used small-intestinal biopsy organ culture to assess CD toxicity that revealed two peptides with selected substitution of both glutamic acid (E) and proline (P) resides abrogated evidence of CD toxicity. Our study suggests the AA deamidated glutamic acid at p65 and proline at p67 are the most important residues in CD toxicity.
In this pilot study, we tested an experimental platform, with which we can systematically investigate variations of gluten molecules with reduced toxicity for CD patients. A critical aspect of this platform is that it provides a screen before proceeding with in vivo challenge in volunteers, the results of which would have significant implications for patient care. Moreover, it allows us to separate CD toxicity from use of gluten for baking purposes. Indeed, review of our prolamin sequencing data(Reference Zhang, Ciclitira and Messing11) suggests there are naturally occurring wheat prolamins that do not contain CD-toxic motifs pertaining to the peptides tested. We propose this could potentially serve as the basis for developing novel treatment for CD. This would involve gluten proteins including gliadins and glutenins that retain the baking qualities of wheat, but which do not exacerbate CD, including α-, γ- and ω-gliadins, low and high molecular weight glutenins, following which the rheology and sensorial characteristics of the resultant flour would need to be assessed.
Acknowledgements
The co-authors wish to thank Ms Janet Schulz, for typing up the manuscript.
N. J., T. S. and B. C.-R. were supported by The Clinical Research Trust; W. Z. and J. M. were supported by the Selman A. Waksman Chair in Molecular Genetics of Rutgers University, NJ, USA, and P. J. C. was supported by Guys and St Thomas NHS Foundation Trust, London, UK.
N. J. with T. S. and B. C.-R. undertook the CD toxicity studies with gluten-sensitive T cells and CD duodenal biopsy organ culture. W. Z. designed and generated the gluten peptides; J. M. supervised W. Z. and was instrumental in design of the study. P. J. C. was jointly instrumental in design of the study and undertook the clinical aspects of the study including taking duodenal biopsies and peripheral blood for purification of antigen presenting cells to generate gluten-sensitive T cells and to undertake CD duodenal biopsy organ culture. He also co-supervised the overall project with J. M. and was instrumental in drafting the manuscript with input from all the co-authors.
The authors declare that the views expressed in the submitted manuscript are their own and not the position of the institution or funders.











