INTRODUCTION
In The Netherlands, over 40 000 cases of gastroenteritis from non-typhoidal Salmonella occur annually [Reference De Wit1]. At least one in six of these cases consults a general practitioner [Reference De Wit2], and diagnostic practices lead to about 2200–3000 laboratory-confirmed cases annually [Reference Van Pelt3, Reference Van Pelt4]. The Dutch laboratory-based Salmonella surveillance covers about 64% of the population, and is based on 16 regional public health laboratories sending all primary Salmonella isolates of patients to the Center for Infectious Disease Control of the National Institute of Public Health and the Environment (RIVM/CIb) for sero- and phage-typing [Reference Van Pelt3]. A sample of Salmonella isolates from food and animals are typed and included in the national surveillance database as well. In this database, a computer algorithm automatically detects an increase in subtypes of human Salmonella isolates, as well as uncommon clusterings by region or age group.
From January 2006 onwards, an increase of a rare phage type of Salmonella Typhimurium, ranging from none to 16 annual cases between 2000 and 2005, was observed. This Dutch phage type 561 (STM561), translates to STM DT7 in the English typing scheme. A request for information to the international surveillance network Enternet showed this type to be rare in Europe as well, only regularly occurring in east Germany. The outbreak accumulated to over 200 laboratory-confirmed cases by the end of the year. Although cases were observed nationwide, more than 55% of the cases originated from a small region in the eastern part of The Netherlands. This large outbreak prompted an investigation, first regionally and subsequently nationally, in order to identify and control the source.
METHODS
Alert and data collection of cases
On 7 February 2006, RIVM/CIb registered an increase of S. Typhimurium phage type DT7 with cases almost exclusively originating from one geographical region designated ‘Twente’ (~616 000 inhabitants). The alert was sent to the responsible municipal public health service (MHS) to investigate the cause of this increase. The alert was accompanied by a routine comprehensive trawling questionnaire and background information about products generally involved in STM cases. Moreover, the MHS was informed about the recent detection of STM DT7 in a pork product taken during routine monitoring from a supermarket in the same region in early February, which was entered into the national Salmonella database. At the MHS a brief questionnaire, targeted at consumption of pork, was transcribed from the trawling questionnaire (Q1, used from 8 February onwards). It was administered to laboratory-confirmed cases. When on 13 March, a sample of cattle manure was found positive for STM DT7 in the Salmonella database, the questionnaire was extended to include beef (products) as well (Q2, from 20 March onwards). The cattle manure sample was found to originate from dairy farm X, in the Twente region. Consequently, dairy products and contact with animals were added to the questionnaire (Q3, from 3 April onwards). Finally, on 18 July the questionnaire was again amended to include additional more detailed questions about recreational activities and consumption of food outside the home (Q4). In all questionnaires, demographic characteristics, clinical manifestation and hospitalization were included. Questions about exposure covered the 7-day interval before onset of illness. Cases were interviewed by telephone.
From 2 July 2006 onwards, all cases with S. Typhimurium cultured at the regional laboratory were reported daily to the MHS, before determination of the phage type, to reduce the time interval between onset of illness and interview of the patients. In July, the MHS dispatched an information leaflet to all general practitioners, paediatricians, and child health centres in the region, to inform them about the outbreak and encourage them to send stool specimens for laboratory testing of all suspected cases in order to increase the sensitivity of the surveillance system.
Case-control study in subset of cases
Based on the questionnaires and a STM DT7-positive manure sample, farm X, a large dairy farm with open access to the public, located in the epicentre of the outbreak, was hypothesized as the source of the outbreak. At this farm, non-pasteurized hard cheese (designated as ‘farmhouse cheese’) was produced (about 500 cheeses produced daily) and sold. On 11 August, it was decided to perform a matched case-control study in order to confirm and specify the vehicle and to prevent further transmission. A case was defined as an individual with gastroenteritis (diarrhoea and at least two additional predefined symptoms) and a laboratory-confirmed STM DT7 infection with date of arrival of the isolate at RIVM/CIb after 1 June 2006. A control was a person registered in the national internet phonebook matched for age group (0–5, 6–10, 11–20, 21–64, ⩾65 years) and 4-digit postcode, and who did not travel abroad in the week preceding the date of onset of the matched case. For each case, two controls were selected. All cases and controls were interviewed by telephone between 14 August and 1 September, using a standardized questionnaire directed at risk exposures to farms, animals and food products. Exposures referred to the 7 days prior to onset of illness for cases, and the week (Monday–Sunday) preceding the date of onset of the matched case for the controls.
Case-case comparison on subset of cases
The immediate reporting of S. Typhimurium cases before determination of the phage type allowed a comparison of data (Q4 questionnaire) for cases with STM DT7 (n=98) vs. cases infected with other phage types (n=39).
Investigation of dairy cattle, cheeserie and dairy products farm X
On 14 April, farm X was visited for hygiene inspection and three samples of hard farmhouse cheese, butter and milk were tested for the presence of Salmonella. For each product a 25-g sample was investigated according to ISO and NEN methods [5–7]. On 19 July, farm X was visited again and 12 samples of hard cheese (25-g samples per cheese) were taken and tested for Salmonella. On 2 August swab samples (n=40) were taken from the shop (products and utensils) and cheese production room. On 9 August, further swabs were taken from the cheese storage room and rinds of various types of cheeses and herbs and spices used in the cheese production were sampled (n=80 samples). After cleaning with steam, the production room was sampled again on 14 August as well as cattle feed, the milk storage room and the petting zoo, in total about 70 samples were taken. All swabs and 25 g of herbs or spices were put in buffered peptone water (BPW; Biotrading, The Netherlands) and incubated for 18–22 h at 37°C. For selective enrichment, modified semi-solid Rappaport–Vassiliadis (MSRV; Difco, USA) plates were inoculated with 100 μl from BPW and incubated for 48 h at 41·5°C. Plates were examined after 24 h and 48 h. Suspected colonies were transferred to Brilliant Green agar (BGA; Oxoid, UK) plates which were incubated for 24 h at 37°C. BGA-suspected colonies were further identified using lysine decarboxylase broth (LDC; Tritium, The Netherlands), triple sugar iron agar (TSI; Tritium) and ureum agar (UA; Tritium). Definitive identification was done by phage-typing and multiple-locus variable-number tandem repeats analysis (MLVA)-typing, as described below.
Through a case interviewed on 24 September 2006, a dairy farm Y, selling cheese produced at farm X, was identified. From eight types of cheese obtained from farm Y on 13 October 2006, 1-kg samples, including the rind, were analysed as described above. One of three Salmonella-positive cheeses had a traceable production date. Ten cheeses from the same production date still present at farm X were received on 4 November 2006, and 0·6 kg of each cheese without the rind was examined as follows. All cheese samples were cut into five pieces of 25 g which were each suspended in 225 ml pre-warmed BPW (45°C), using a stomacher for 30 s at normal speed. Stomacher bags were incubated 24 h at 37°C. For selective enrichment, we used MSRV plates (see above), Rappaport–Vassiliadis soya peptone broth (RVS, Oxoid) and Muller–Kaufmann Tetrathionate broth, supplemented with novobiocine (MKTTn; Oxoid). RVS and MKTTn were incubated for 48 h at 37°C. After 24 h and 48 h, the possible presence of Salmonella was checked by streaking a small amount of sample from MSRV, RVS and MKTTn on xylose, lysine, Na-desoxycholate agar (XLD, Oxoid) and BGA plates, which were incubated for 24 h at 37°C. Suspected colonies were further identified using LDC, TSI and UA.
Finally, two complete cheeses from the contaminated batch were examined. The cheeses were cut into nine segments of about 1 kg each. From each kilogram, 5×25 g, 5×2·5 g and 5×0·25 g were examined (without rind). Using the tables from DeMan [Reference DeMan8], the most probable number of salmonella present in these cheeses was determined.
Since June 2000, farm X had participated in a voluntary Salmonella certification programme of bulk milk run by the Animal Health Service. In this programme, the presence of antibodies to Salmonella in bulk milk is tested up to three times a year using a mixed-LPS ELISA for Salmonella groups B and D, which is an extension of the D-LPS ELISA described by Veling et al. [Reference Veling9]. The Salmonella status of the dairy cattle from farm X was obtained for 2005 (the year with the first Salmonella-positive test result) and 2006. Further, the Animal Health Service intensified their investigations on farm X following the ‘Salmonella suspect’ status and reported the results retrospectively.
Laboratory investigation: typing of isolates
The regional Public Health Laboratory for Medical Microbiology Twente Achterhoek sends all primary Salmonella isolates of patients to the National Reference Laboratory for Salmonella. Isolates are routinely serotyped, and S. Enteritidis and S. Typhimurium are additionally phage-typed [Reference Van Duijkeren10]. For further discrimination within phage types, isolates obtained during the outbreak investigation were additionally typed by pulsed-field gel electrophoresis (PFGE) (data not shown) [Reference Gatto11] and MLVA [Reference Lindstedt12], with slight modifications according to Torpdahl et al. [Reference Torpdahl13].
Statistical analysis
Descriptive analyses were performed to assess frequency of clinical symptoms, hospitalization rate and frequencies of exposure. In the case-case comparison, associations between an infection with STM DT7 and exposures were assessed by odds ratios (OR) and 95% confidence intervals (CI). In the case-control study, associations between infection and exposures were assessed by OR and 95% CI obtained by conditional logistic regression (SAS 9.1.3, SAS Institute Inc., USA). Matching strata consisted of the individual strata of each case and the corresponding controls. Gender and factors associated with disease in bivariate analyses with a P value ⩽0·20 were included in the multivariate analyses. The attributable fraction in the total number of cases in the case-control study was simply calculated as the proportion of cases reporting the risk factor(s).
RESULTS
Descriptive analysis of the outbreak cases
In total, 224 laboratory-confirmed cases with STM DT7 were identified between 1 January 2006 and 15 April 2007 (Fig. 1). Of these, 129 (58%) occurred in the Twente district and 51% were male. Of all cases, 44% were aged 0–5 years, 18% 6–10 years, 16% 11–20 years, 14% 21–64 years and 8% ⩾65 years (median 7·7 years, range 0–93 years).
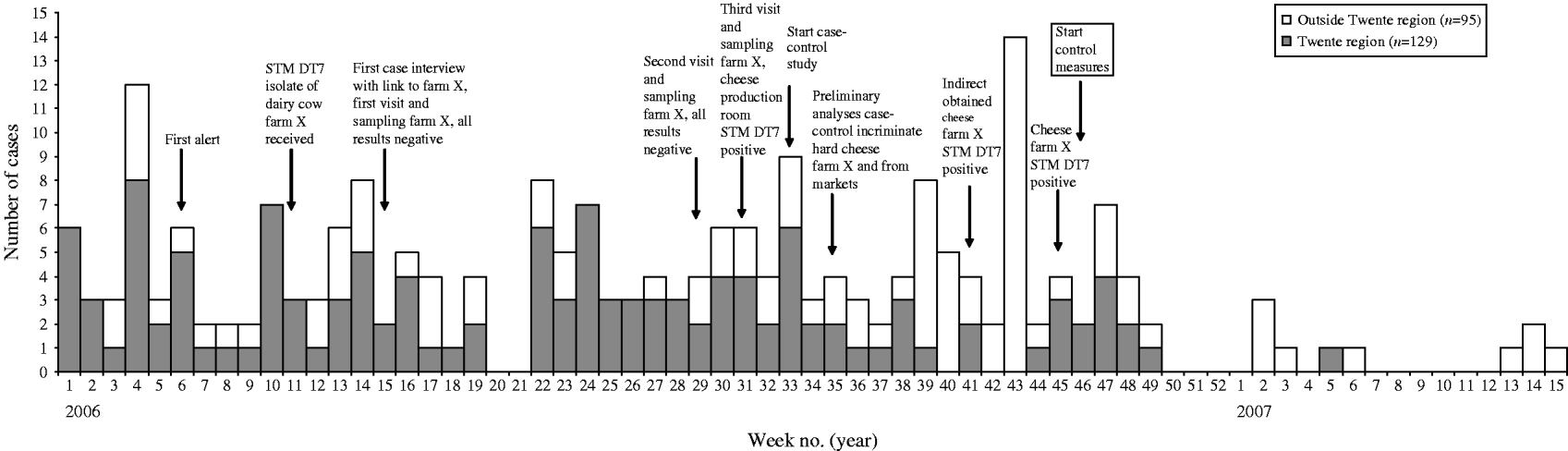
Fig. 1. Laboratory-confirmed cases with Salmonella Typhimurium phage type 561 (STM DT7) infection (n=224) by week of isolate registration at RIVM, The Netherlands, 1 January 2006 to 15 April 2007.
For 175 (78%) of the cases, questionnaire data (Q1–Q4) were available. These included 109 cases from the Twente district (84% of all cases in this district) and 66 from outside this district (69% of cases outside the district). Most common symptoms were diarrhoea (98%), fever (76%), blood in stool (68%) and vomiting (47%). Of the cases, 59 (35%) were hospitalized, with a median stay of 4 days (range 1–41 days).
Case-control study of subset of cases
In the case-control study, 51 cases and 105 matched controls were included. Of the 51 matched sets, 37 (73%) were resident in the Twente district. Risk factors univariately associated with disease with a P value <0·20 were: visit to a farm; or consumed or bought hard cheese, herb cheese, milk, butter milk and yoghurt from a farm; or consumed or bought dairy products in general and hard cheese specifically from a market. Conditional multiple logistic regression analyses indicated that cheese purchased from a farm, and specifically from dairy farm X and from a market stall, were associated with infection with STM DT7 (Table 1). Overall, 27 (53%) of the cases in this study reported one of these two risk factors.
Table 1. Risk factors identified by multiple conditional logistic regression analysis, case-control study, 51 cases and 105 controls, The Netherlands, 2006
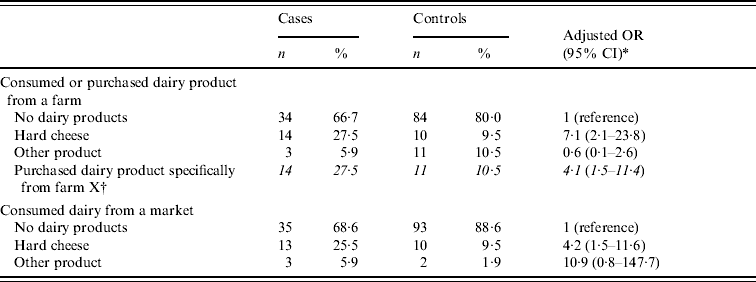
OR, Odds ratio; CI, confidence interval.
* Also adjusted for gender included in the final model.
† Derived from sub-analyses with different response categories for variable ‘Consumed or purchased dairy product from a farm: no/yes from farm X, yes from other farm. All positive responses for farm X refer to hard cheese, except for one control. No other specific farms were associated with STM DT7.
Case-case comparison
Of the 98 STM DT7 cases in the case-case comparison, 29 (30%) had bought dairy products, mainly hard cheese, produced at farm X, compared to none of the 39 cases with a different phage type of S. Typhimurium (OR=infinite).
Cases explained by incriminated dairy farm products
In total, for 133 cases (59%) information on exposure to the incriminated farm products was available from questionnaires Q3 and Q4. For 42 (31%) of these STM DT7 cases a direct link to the specific dairy farm X was reported. In addition, 40 cases had consumed hard farmhouse cheese (38) or other dairy farm products (2), originating from other or unknown sources.
Investigation of dairy cattle, cheeserie and dairy products from farm X
All 25-g samples of the dairy products sampled in April and July 2006 tested negative for STM DT7. Following the visits on 2 and 9 August, STM DT7 was detected in the drain and from a cheese transport vehicle, both in the cheese production room. All samples (cheese, cattle feed, petting zoo) taken on 14 August tested negative. In October, from the eight types of cheese produced by farm X obtained from farm Y, three were found positive (i.e. at least one of the five 25-g samples per cheese was positive): a 6-week-old cheese, a 9-month-old cheese and a mustard seed cheese of unknown age. Only for the 9-month-old cheese, could the production date be traced. From ten cheeses of the same production date obtained from farm X, two were positive for STM DT7: in cheese 1, one of five 25-g samples positive; in cheese 2, all five 25-g samples positive. Of the two complete cheeses from the contaminated batch that were subsequently examined, one cheese was found negative and the second appeared to be contaminated with a most probable number of 4·2 salmonella/kg (95% CI 1·5–9·1 c.f.u/kg).
Results from the voluntary Salmonella certification programme of the Animal Health Service showed that the bulk milk on farm X tested positive for the first time in January 2005, indicating that at least 10% of the milk-producing cows were serologically positive for Salmonella (groups B or D). Bulk milk tested positive at the revisit in December 2005, demonstrating persistent infections. No clinical disease was observed in the dairy cattle in 2005. Subsequent bacteriological investigation of three dung pits in February 2006 demonstrated the presence of Salmonella in two of them (not typed further). Repeated testing of the dung pits on 15 August, 18 October and 29 November 2006 showed no positive results. In March 2006, one dairy cow developed diarrhoea and the manure tested positive for Salmonella, typed as STM DT7 and highlighted in the national database. This was the first link between the outbreak and farm X. No further symptoms were observed in the cattle in 2006. The cattle herd (those aged ⩾6 months) was investigated for Salmonella three times during 2006 to identify Salmonella carriers. During the first study, in March–June, 29 cows were repeatedly found serologically positive and another four animals excreted Salmonella in manure. The four excreters were slaughtered. In September–November 20 cows were serologically positive again and were slaughtered subsequently, but no excreter cows were found. Finally, on 23 November, 12 cows were found serologically positive for the first time. The farmer did not wait to see if these infections were persistent and all 12 were slaughtered immediately. The positive results of the voluntary Salmonella certification programme in 2005 and February 2006 were not communicated to the MHS or Food and Consumer Product Safety Authority because of privacy legislation.
Laboratory investigation: typing of isolates
As a national phage-typing scheme is used for S. Typhimurium, 47 STM561 isolates were also typed using the international phage-typing scheme, and were found to correspond with DT7. For 213 (95·1%) of the 224 cases, MLVA results were available (Fig. 2). Overall, 22 different MLVA types were observed, of which three types represented 81·3% of all isolates: the main type was 02-06-05-00-02 (60·1%), followed by the related type (one locus difference) 02-06-04-00-02 (12·7%) and another related type 02-06-05-00-00 (8·5%). Other related MLVA types were found for 22 additional isolates (10·3%). The remaining 18 isolates differed by more than one locus from the main MLVA type. Of 42 cases with known exposure to the incriminated dairy farm (products), 29 cases (69%) were infected with the main type, nine with related type 02-06-05-00-00 (21%), three with other related types and one with a strain that had more than one locus difference.
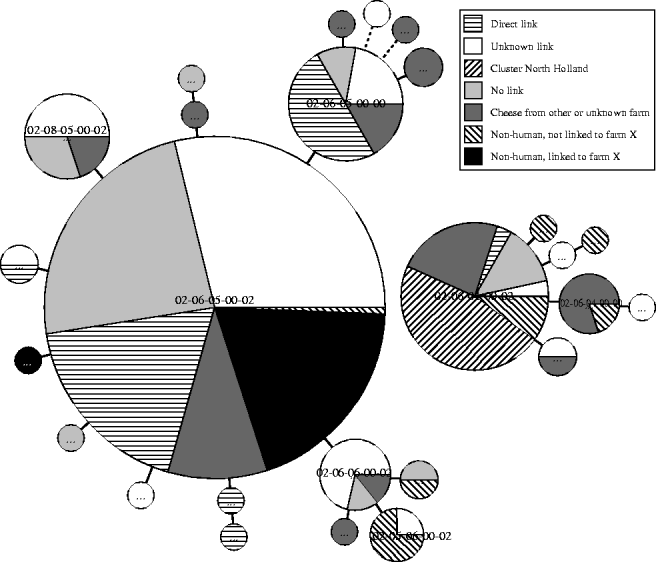
Fig. 2. MLVA results of STM DT7 isolates of 213 cases and 43 STM DT7 isolates of non-human origin, 1 January 2006 to 15 April 2007.
Control measures
When on 14 November 2006, the cheese was confirmed to be contaminated by STM DT7, several control measures were implemented by COKZ, the primary supervisor of dairy production for approved premises in The Netherlands. All cheeses with production dates between 1 November 2004 and 30 April 2006 still available at the farm or at two large wholesalers were immediately destroyed or melted for melt-cheese spread (>350 cheeses). Subsequently, a daily production sample of cheeses produced since 1 May 2006 was tested for the presence of Salmonella, and if positive was destroyed (total number of destroyed cheeses unknown, at least 100). Furthermore, an extensive plan for improved hygiene in the production process was implemented, including a professional advisor for detection and elimination of contamination routes at the farm and control on milk temperature and growth of enterobacteriaceae. From the end of November 2006 until 22 January 2007, the milk for cheese production was pasteurized. During 2007, production of unpasteurized hard farmhouse cheese was reintroduced under close supervision of COKZ, checking samples of each weekly production for Salmonella. No Salmonella was observed in the cheese after implementation of the control measures.
DISCUSSION
The case-control study combined with microbiological detection of STM DT7 in dairy cattle, the cheese production room and finally cheeses from a dairy farm, incriminated hard cheese made from unpasteurized milk as the source of the nationwide STM DT7 outbreak. The outbreak was identified following routine surveillance, including sero- and phage-typing. It was only due to the unusual phage type that the outbreak was noticed. The integration of human, veterinary and food data into a common Salmonella database is considered crucial for early warning and source identification. Since only a minority of gastroenteritis patients seek medical advice and only some of these become laboratory-confirmed cases, the real number of affected individuals in this outbreak was clearly higher, estimated at about 3000 cases.
Generally, cheese-associated Salmonella outbreaks can be attributed to improper or no pasteurization or post-pasteurization contamination and are mainly related to consumption of soft cheeses, such as Mexican-style, raw-milk queso fresco, mozzarella, fresh Cantal cheese and Brie or Camembert [Reference Djuretic, Wall and Nichols14–Reference Austin21]. Hard cheese made from unpasteurized milk is an uncommon source of Salmonella infections, although outbreaks due to cheddar are occasionally described [Reference Djuretic, Wall and Nichols14, Reference Altekruse15, Reference Fontaine22]. Hard cheese from unpasteurized milk is legally sold as farmhouse cheese in The Netherlands. This outbreak demonstrates that hard, unpasteurized cheeses, even aged varieties (9 months), should not be excluded as a possible vehicle for Salmonella infections. The long shelf-life of hard aged cheeses, ⩾12 months for fully mature cheese, explains the protracted course of the outbreak and makes it possible for successful intervention, and prevention of further cases.
Quantitative analysis of two whole cheeses showed that contamination could be as low as 4·2 salmonella/kg (95% CI 1·5–9·1). Most samples found positive for Salmonella in our investigation contained a number that was within this 95% CI. As we examined a limited number of cheeses, this number might not be representative of all contaminated cheeses involved in the outbreak. However, in previous (soft) cheese outbreaks, similar low-level contamination was observed [Reference Fontaine22–Reference Hedberg24]. This would explain why Salmonella was not detected by the Food and Consumer Product Safety Authority following the farm visits in April and July 2006, by routinely testing a 25-g sample of each cheese. According to the Commission Regulation (EC) No. 2073/2005 on microbiological criteria for foodstuffs, which applied from 1 January 2006, for cheese, butter and cream produced from raw milk, five samples of 25 g should be tested, and none should be positive [25]. This was not yet implemented in The Netherlands at the time of the investigation. This outbreak clearly supports the scientific base for this criterion. An intensive sampling scheme seems especially important for suspicious food products with a high fat content and mildly acidic pH, both present in cheese. Salmonella in these circumstances/matrices can pass the barrier in the human stomach (the gastric acid), and thus cause disease in many individuals, even if the contamination of the product is low. This might also apply for products such as chocolate and peanut butter that have caused major outbreaks [Reference Kapperud26–Reference Kirk29]. In addition to bacteriological quality monitoring of the end product, hazard analysis critical control point procedures should, of course, be implemented.
A timely and adequate response was hampered in this outbreak. Control measures were not implemented for 9 months. This was mainly due to lack of prompt information exchange between involved human and food safety and veterinary partners, a data collection strategy that started too slowly and initially was limited to meat only, and difficulties in detection of the pathogen in the cheese. This outbreak clearly illustrates that it is ill-advised to target certain food products because similar, even rare, subtypes were detected in the products. It is strongly recommended using comprehensive trawling questionnaires addressing all possible sources and/or open-ended interviews in the initial stage of the investigation. From a public health perspective, earlier measures were clearly needed. Within 2 months following the alert, strong circumstantial evidence was available, such as a highly unique Salmonella phage type isolated from dairy cattle at farm X and the mention of the dairy farm during interviews of some of the early cases. Six months after the alert, the farm was strongly implicated when the same rare Salmonella type was detected in the cheeserie and when in month seven the case-control study results were available. The EU General Food Law (EC decision 178/2002) offers the opportunity to recall food if there are reasonable grounds to suspect that a food product is unsafe, even if the microbiological food safety criteria are met. So far, this precautionary principle has hardly been used in The Netherlands. As a result of this outbreak, a task force was established to define criteria for its use in the future. Although obviously late, eventually the control measures still prevented plenty cases as many cheeses were recalled.
The attributive fraction for proven consumption of dairy products or direct animal contact from farm X may seem relatively low (31%). However, many cases might have consumed contaminated cheese from this particular farm via other cheese sale outlets, who purchased the cheese either directly from the farm (in the region) or from two main wholesalers (nationwide). Only in late November 2006, did the lists of buying shops from these wholesalers become available. These showed that 125 different sales outlets had bought in total at least 2442 cheeses produced at farm X in weeks 1–34 in 2006. At least 156 (6%) of these cheeses were still in stock at one wholesaler and were destroyed. The number of cheeses destroyed by the second wholesaler is unknown. Since for many cases information on places where they had purchased their cheese in the risk period was limited, it was not possible to assess all cases exposed to the contaminated cheese retrospectively. Consequently, true exposure to cheese from farm X was underestimated by the available questionnaire data.
As a fairly new typing tool, the value of MLVA in outbreak investigations of S. Typhimurium needs thorough evaluation. In Scandinavian countries, it was concluded that MLVA was superior to PFGE for both surveillance and outbreak investigations, and particularly valuable for discriminating within the most common phage types, such as DT12, DT104 and DT120 [Reference Torpdahl13, Reference Nygard30, Reference Ethelberg31]. In our study, MLVA results demonstrated a high variability of the human strains within the same rare phage type. Of the human strains epidemiologically linked to the incriminated farm by the questionnaires, 98% had either the main (69%) or a related (29%) subtype. This suggests that a one-locus difference in the MLVA of S. Typhimurium does not necessarily point to a different source of infection and should not lead to exclusion of cases from an outbreak investigation. For a cluster of 14 cases (6%) from a geographically small, distinct area in the western part of the country, contaminated minced meat was the likely source according to the epidemiological information, and no link could be identified with farmhouse cheese [Reference Zwart32]. Nevertheless, MLVA for this cluster detected a subtype related to the main type which, however, was found only once in the cases epidemiologically linked to the farmhouse cheese. It is clear that for valid linkage of cases to an outbreak and to food, both epidemiological and subtyping data need to be considered together.
In conclusion, contaminated hard farmhouse cheese was found to be the vehicle of a large-scale Salmonella outbreak in The Netherlands in 2006. This outbreak shows that hard aged raw-milk cheeses are often undeservedly considered microbiologically safe. A combination of both microbiological and epidemiological investigations and exchange of information, jointly by veterinary, food safety and public health partners, is the most powerful tool for linking cases of foodborne zoonoses and identification of the source. Individuals at increased risk for (severe) Salmonella infections, such as young children, immunocompromised persons, pregnant women and the elderly, should be strongly advised to avoid raw milk products. As a prerequisite, correct labelling, not only at retail, but also in delicatessen counters and shops, should be enforced.
ACKNOWLEDGEMENTS
The Outbreak Investigation Team acknowledge several colleagues of the Epidemiology Department of the RIVM, especially Alies van Lier, Anita Suijkerbuijk, and Anneke Westerhof for help in interviewing cases and controls; Henny Maas, Anjo Verbruggen and Frans Bensink for help in phenotyping of the Salmonella isolates, Mona van Spijk, from the LTO, Noord for advice during the investigation, Antoon Mentink from The Netherlands controlling authority for milk and milk products for his efforts and input on the control measures and Dr Lindstedt of the Norwegian Institute of Public Health for MLVA-typing some of the Salmonella isolates early in the outbreak. Finally, we thank the Enternet participants for their response to the request for action and specifically Wolfgang Rabsch of the Wernigerode Branch of the Robert Koch Institute for sharing some German S. Typhimurium DT7 strains with our laboratory. The work was within the routine activities financially supported by the Ministry of Public Health, Welfare and Sports and local public health funds.
APPENDIX
Remaining members of Outbreak Investigation Team
Public Health Service Regio Twente: Barbara Beuvink, Nettie Schuurman, Anja Brunner, Anjes Timmerman, Jildou Pal, Jan Roorda. Laboratory for Medical Microbiology Twente Achterhoek: Ellanie Koldeweid, RIVM/CIb: Iris van Ouwerkerk, Carolien de Jager, Yvonne Doorduyn, Frans van Leusden, Martijn van Luit, Max Heck. Food and Consumer Product Safety Authority: Mello Sevinga, Albert Lam. Animal Health Service: Jan Veling.
DECLARATION OF INTEREST
None.





