Escherichia coli sequence type (ST131) is a recently emerged, multidrug-resistant lineage of E. coli that has expanded tremendously over the past two decades to become the main cause of extraintestinal E. coli infections in humans [Reference Stoesser1, Reference Nicolas-Chanoine, Bertrand and Madec2]. Within ST131, the H30R subclone predominates and accounts for almost all fluoroquinolone-resistant isolates [Reference Banerjee3]. Within ST131-H30R, the H30Rx subclone is additionally associated with the CTX-M-15 extended-spectrum β-lactamase, conferring resistance to extended-spectrum cephalosporins. This broad and often unanticipated resistance leads to adverse clinical outcomes [Reference Can4].
Preventive interventions against these threatening new pathogens are sorely needed, but few modifiable risk factors have been discovered. Many studies have identified clinical and healthcare-associated risk factors for ST131 infection, usually based on data obtained by medical record review. Here, among US military veterans, we sought novel risk factors for having an ST131-H30 clinical isolate by investigating self-reported clinical factors, demographic characteristics and environmental exposures.
Methods
Study subjects and E. coli isolates
Consecutive clinical E. coli isolates were collected from the Minneapolis VA Medical Center (MVAMC; Minneapolis, MN) clinical microbiology laboratory from May 2012 to December 2013. The source patients were sent an invitation to participate in the study and a paper-based questionnaire, which requested demographic and exposure information. Participants returned completed questionnaires by mail. Telephone follow-up was used to facilitate survey completion for some subjects, without knowledge of laboratory testing results. The MVAMC institutional review board (4181-A) approved the study.
Clinical microbiology
The MVAMC clinical laboratory identified the E. coli isolates using standard methods and determined susceptibility to 12 antimicrobial agents and extended-spectrum beta-lactamase (ESBL) production using the BD Phoenix Automated Microbiology System (Becton Dickinson and Company, Sparks, MD, USA) per manufacturer guidelines, utilising NMIC/ID-123 panels. Minimum inhibitory concentration (MIC) results were interpreted according to Clinical Laboratory and Standards Institute criteria [5].
Molecular typing
Isolates were characterised using established polymerse chain reaction (PCR)-based assays for membership in the ST131-H30 clonal subset or its ST131-H30Rx sublineage [Reference Banerjee3] and O type (O16 and O25b only) [Reference Johnson6]. PCR was done using boiled lysates for template DNA, with appropriate positive and negative control strains.
Statistical analysis
Comparisons of proportions were made using χ 2 and Fisher exact tests. Exposures associated significantly with either ST131 or H30 were included in multiple logistic regression models, which were performed separately for clinical and environmental risk factors.
Results
Clinical and epidemiological data
Over the 20-month study period (May 2012–December 2013), the MVAMC clinical microbiology laboratory recovered E. coli from 925 unique veterans, of whom 469 (51%) opted to complete the study survey. Most subjects were male (82%); half were 66 years or older (Table 1). Most were seen in the outpatient setting (384; 83%); the rest (79; 17%) had been hospitalised for 48 h or more. The most common specimen types were urine (366; 78%), wound swab (52; 11%) and blood (15; 3%).
Table 1. Characteristics of 469 veterans with an Escherichia coli clinical isolate, stratified by ST131 status

All variables included results for between 462 and 469 subjects.
a Subjects could report more than one co-morbidity.
*P-value corresponds to an overall χ 2 test.
Antimicrobial susceptibility
Ampicillin was the most commonly detected resistance phenotype (187; 40%), followed by ciprofloxacin (84; 18%), ampicillin-sulbactam (80; 17%) and trimethoprim-sulfamethoxazole (77; 16%). No isolate was carbapenem-resistant. Eighty-five isolates (18%) were resistant to ⩾3 antimicrobial agents and 21 (4%) to ⩾5 antimicrobial agents.
Prevalence of ST131 and its subsets
ST131 accounted for 66 (14%) of the 469 study isolates. Of the 66 ST131 isolates, 51 (77%) were ST131-H30, of which 43 (84%) represented the H30R1 subclone and 8 (12%) the ST131-H30Rx subclone. Subjects with ST131 vs. non-ST131 isolates did not differ significantly on demographic characteristics, including age, dwelling type, health status, or comorbidities (Table 1).
Among the 66 ST131 isolates, all 51 H30 isolates, but only 2/15 (13%) non-H30 isolates, were fluoroquinolone-resistant (P-value < 0.001). Sixty-three (95%) of the ST131 isolates were typed as O25b; 51 (88%) of these were H30.
Clinical factors and ST131 and ST131-H30
Overall, ST131 was significantly associated with exposure to medical lines (within the past month (P-value = 0.005) or 6 months (P-value < 0.001)), hospitalisation (P-value = 0.004), nursing home residence (P-value = 0.001), surgery (P-value = 0.01) and antimicrobial exposure in the past 6 months (P-value = 0.008) (Table 2). ST131 also was over-represented among isolates from non-blood, wound, or urine specimens (P-value = 0.01) (Table 2).
Table 2. Clinical factors and E. coli ST131 carriage, by ciprofloxacin resistance and ST131 H30 sub-group
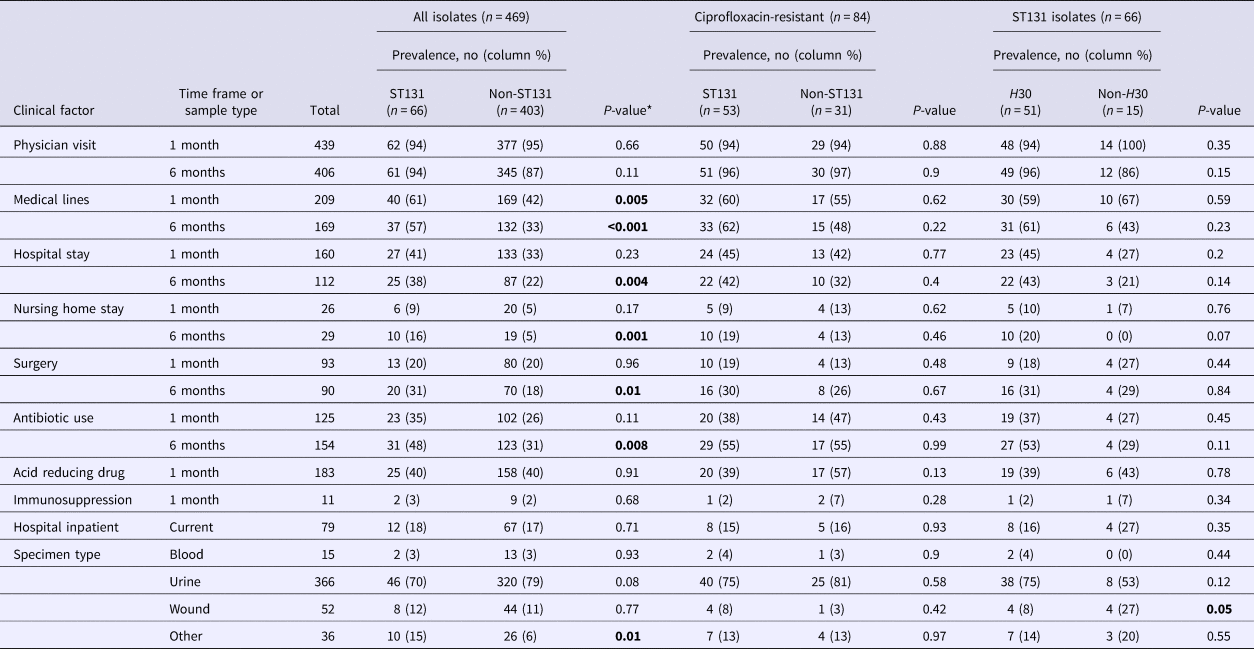
*For some variables data were missing for 2–3 subjects. Hypothesis testing with small numbers (cell size <5) was conducted using Fisher's exact test, otherwise, the P-value for the χ 2 test is reported. Bold P-values reflect statistically significant differences.
By contrast, among ciprofloxacin-resistant isolates, clinical risk factors did not differ significantly between ST131 (n = 53) and non-ST131 (n = 31) isolates (Table 2). Similarly, among ST131 isolates, the only clinical risk factor that differed H30 from non-H30 isolates was wound source, which was weakly associated with non-H30 isolates (Table 2).
Multiple logistic regression was used to identify clinical variables independently associated with E. coli ST131. Overall, after adjustment for variables significantly associated with ST131 in the univariable analyses, only medical line used in the past 6 months (adjusted OR 2.14, 95% CI 1.23–3.74) and nursing home residence (adjusted OR 2.92, 95% CI 1.3–6.76) remained significantly associated with ST131. In univariable analyses, no clinical variable was associated with ST131 status among ciprofloxacin-resistant isolates and only one variable (wound source) was associated with H30 status among ST131 isolates (Table 2) and for these subsets, multiple logistic regression was not performed.
Environmental factors and ST131 and ST131-H30
Overall, ST131 was associated positively with touching wild animals (P-value = 0.03) and negatively with meat consumption (P-value = 0.01), cat contact (P-value = 0.02) and touching animal feces (P-value = 0.03) (Table 3). Among subjects with a ciprofloxacin-resistant isolate (n = 84), ST131 was associated positively with farm animal contact (P-value = 0.05) and touching wild animals (P-value = 0.02) and negatively with using city water (P-value = 0.05) and cat contact (P-value = 0.01). Among subjects with an ST131 isolate (n = 66), H30 was associated negatively with past-month out-of-state travel (P-value = 0.05).
Table 3. Environmental factors and Escherichia coli ST131 carriage, by ciprofloxacin resistance and ST131-H30 sub-group
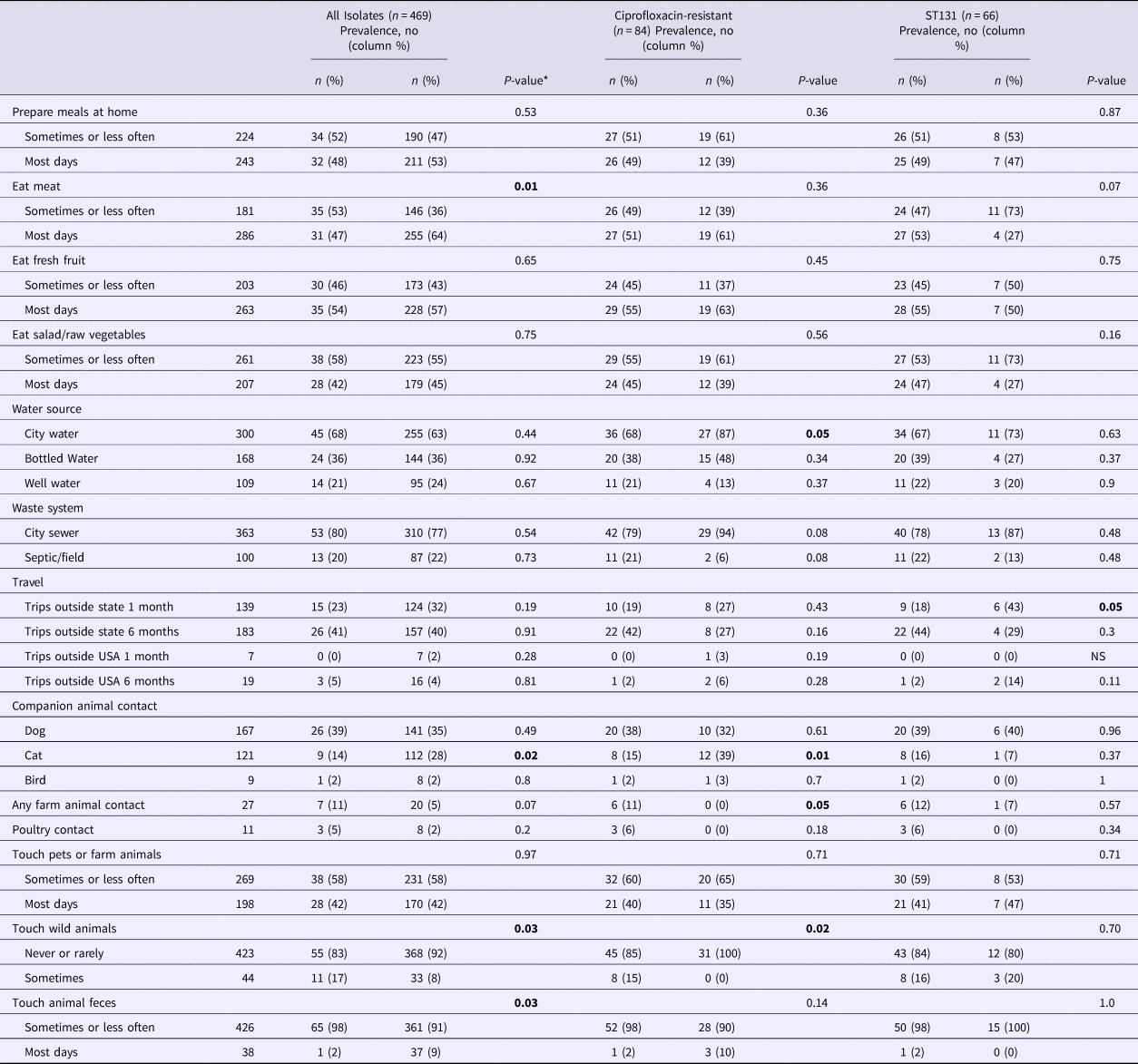
*For some variables, data were missing for 2–3 subjects. Hypothesis testing with small numbers (cell size <5) was conducted using Fisher's exact test, otherwise, the P-value for the χ 2 test is reported. Bold P-values reflect statistically significant differences.
Multiple logistic regression was used to identify environmental variables independently associated with carriage of E. coli ST131. After adjustment for variables associated with ST131 in the univariable analyses, contact with wild animals (adjusted OR 2.49, 95% CI 1.16–5.33) remained positively associated and meat consumption (adjusted OR 0.48, 95% CI 0.28–0.82) and pet cat exposure (adjusted OR 0.37, 95% CI 0.17–0.77) remained negatively associated, with ST131. With wild animal contact in the model, contact with animal feces was no longer associated with ST131 (data not shown). Multiple logistic regression was not performed for subjects with ciprofloxacin-resistant E. coli, all of whom reported contact with wild or companion animals, or for subjects with ST131 since within this subset only one H30-associated variable (out-of-state travel) was identified.
Discussion
Here we assessed self-reported demographics, clinical factors and environmental exposures of veterans with an E. coli clinical isolate as predictors of ST131 status (both overall and for fluoroquinolone-resistant isolates) and, among subjects with an ST131 isolate, of H30 subclone status. This is one of few studies to examine non-healthcare-associated risk factors for having a clinical isolate from the pandemic ST131 lineage and specifically its H30 subclone.
Our findings support two main conclusions. First, most of the ST131-associated clinical factors represent traditionally recognised risk factors for having an antimicrobial-resistant infection. However, with stratification by fluoroquinolone resistance status, the ST131-specific associations disappeared. This suggests that these healthcare exposures are not specific to ST131, but are generic markers for infection with antimicrobial-resistant E. coli generally.
Second, a modestly (albeit statistically significantly) greater proportion of subjects with non-ST131 E. coli, as opposed to ST131, reported frequent exposure to certain hypothesised environmental risk factors. Specifically, non-ST131 group members reported more frequent meat consumption, cat ownership, city water usage, recent out-of-state travel and touching animal feces. By contrast, ST131 group members reported more contact with wild animals. This suggests that contact with wild animals, or other variables associated with wild animal exposure, predispose specifically to having ST131, as has been suggested by others [Reference Müller, Stephan and Nüesch-Inderbinen7]. Carriage of the ST131-H30 subclone was associated with wound specimens, while non-H30 carriage was associated with past-month out-of-state travel.
The study has several limitations. First, relatively few subjects reported having several of the candidate risk factors (e.g. travel or farm animal exposure), or had ST131-H30Rx, limiting statistical power for these variables. Second, all subjects were veterans, limiting generalisability and possibly contributing to our not finding the typical associations of ST131 with older age, comorbidities and overall poor health [Reference Blanco8–Reference Price, Johnson and Aziz10]. Third, the requirement for survey completion may have selected for an uncharacteristically healthy and functional subset of veterans. Fourth, risk factors for E. coli acquisition or carriage may have occurred earlier in life and hence were unmeasured in this study. Additional limitations include the risks of false positives from multiple testing and of misclassification due to poor subject recall.
Specific pandemic lineages clearly contribute disproportionately to extraintestinal E. coli infections, especially if antimicrobial-resistant, making their potential reservoirs and modes of transmission important to understand. In this veteran-based study population, ST131 was associated preferentially with healthcare-associated exposures and wild animal contact, whereas non-ST131 E. coli were associated with other environmental exposures.
Author ORCIDs
Amee R. Manges 0000-0003-2462-7249.
Acknowledgements
This work was supported in part by Office of Research and Development, Department of Veterans Affairs (USA), grants 1 I01 CX000920-01A1 and 2 I01CX000920-04 (to. J.R.J.). The views represented here are those of the authors and not those of their respective institutions or the Department of Veterans Affairs.
Conflict of interest
Dr Johnson has received research funding from Merck, Allergan and Tetraphase, is a consultant for Janssen/Crucell and has patent applications for tests to detect E. coli strains. The other authors report no financial conflicts of interest.





