Endometriosis is the most common cause of pelvic pain that affects 6–10% of women in their reproductive years (Treloar et al., Reference Treloar, O'Connor, O'Connor and Martin1999) and 20–50% of women with infertility (Gao et al., Reference Gao, Outley, Botteman, Spalding, Simon and Pashos2006). It is primarily characterized by the presence of endometrium-like tissue in cavities other than the uterus. Besides severe pelvic pain, women with endometriosis may also suffer from heavy or irregular menstrual bleeding, pain during intercourse and exercise, infertility, lower abdominal and back pain, diarrhea, and/or constipation and chronic fatigue. Endometriosis has a complex etiology that results from an interplay of genetic and non-genetic risk factors and has an estimated heritability of approximately 51% (Treloar et al., Reference Treloar, O'Connor, O'Connor and Martin1999).
We and others have conducted genome-wide association (GWA) studies involving individuals of European and Japanese ancestries to identify genetic risk factors for endometriosis (Albertsen et al., Reference Albertsen, Chettier, Farrington and Ward2013; Nyholt et al., Reference Nyholt, Low, Anderson, Painter, Uno, Morris and Montgomery2012; Painter et al., Reference Painter, Anderson, Nyholt, Macgregor, Lin, Lee and Zondervan2011; Uno et al., Reference Uno, Zembutsu, Hirasawa, Takahashi, Kubo, Akahane and Nakamura2010). Results from the four GWA studies strongly associated nine independent single nucleotide polymorphism (SNP) loci with endometriosis, providing genome-wide significant (p < 5 × 10−8) evidence in at least one study. These risk loci include the following: rs10965235 in the CDKN2BAS gene at 9p21.3, rs12700667 at 7p15.2, rs7521902 near WNT4 at 1p36.12, rs13391619 in the GREB1 at 2p25.1, rs10859871 near VEZT at 12q22, rs4141819 at 2p14, rs7739264 near ID4 at 6p22.3, rs1537377 near CDKN2B-AS1 at 9p21.3, and rs1519761 at 2q23.3. The GREB1 signal was implicated in a meta-analysis of the European (QIMRHCS+OX) and Japanese (BBJ) GWA data after combining published results for rs1339416 from Adachi et al. (Reference Adachi, Tajima, Quan, Haino, Yoshihara, Masuzaki and Tanaka2010) — a small GWA study comprising 696 endometriosis cases and 825 controls of Japanese descent. A recent follow-up study by ourselves further implicated GREB1 locus in endometriosis risk, with stronger association signals within the region (Fung et al., Reference Fung, Holdsworth-Carson, Sapkota, Zhao, Jones, Girling and Montgomery2015). Three risk loci (rs4141819 at 2p14, rs7739264 near ID4 at 6p22.3, and rs1537377 near CDKN2B-AS1 at 9p21.3) were implicated when analysis was conducted after excluding endometriosis cases with minimal or mild (revised American Fertility Society [rAFS] stage 1 or 2 disease; American Society for Reproductive Medicine, 1997) endometriosis (Nyholt et al., Reference Nyholt, Low, Anderson, Painter, Uno, Morris and Montgomery2012). More recently, we conducted a candidate-gene association study (Sapkota, et al., Reference Sapkota, Low, Attia, Gordon, Henders, Holliday and Nyholt2015b) to investigate potential role of the interleukin 1A (IL1A) variants reported by two small Japanese GWA studies for endometriosis (Adachi et al., Reference Adachi, Tajima, Quan, Haino, Yoshihara, Masuzaki and Tanaka2010; Hata et al., Reference Hata, Nakaoka, Yoshihara, Adachi, Haino, Yamaguchi and Tanaka2013). The study results provided genome-wide significant evidence for the association of rs6542095 in IL1A with endometriosis. Taken together, results suggest 10 independent SNP loci for endometriosis at genome-wide significant level; of these, nine are polymorphic in populations of European ancestry.
The three replication studies (Pagliardini et al., Reference Pagliardini, Gentilini, Vigano, Panina-Bordignon, Busacca, Candiani and Di Blasio2013, Reference Pagliardini, Gentilini, Sanchez, Candiani, Vigano and Di Blasio2015; Sundqvist et al., Reference Sundqvist, Xu, Vodolazkaia, Fassbender, Kyama, Bokor and Falconer2013) performed to date have only replicated association of rs7521902 (p = 5.6 × 10−3; Pagliardini et al., Reference Pagliardini, Gentilini, Vigano, Panina-Bordignon, Busacca, Candiani and Di Blasio2013) and rs10859871 (p = 6.9 × 10−5; Pagliardini et al., Reference Pagliardini, Gentilini, Sanchez, Candiani, Vigano and Di Blasio2015) with endometriosis. Inherently hidden fine stratification among different populations of similar ancestry, which is often difficult to tease apart, power, and possible variations in disease definition and/or classification may have contributed to these inconsistent results. Moreover, these replication studies have investigated only a fraction of the implicated SNP loci, and hence additional replication studies are required to examine association of all risk loci with endometriosis.
Here we extended our previous study (Sundqvist et al., Reference Sundqvist, Xu, Vodolazkaia, Fassbender, Kyama, Bokor and Falconer2013) to evaluate all nine implicated SNP loci for endometriosis that are polymorphic in populations of European ancestry, utilizing GWA data for surgically confirmed 998 endometriosis cases and 783 disease-free controls from Belgium. Importantly, severity of disease in endometriosis cases has been prospectively graded using the rAFS classification system, and hence is less likely to be misclassified as compared with the retrospective disease staging based on clinical records. We also performed meta-analysis for the nine risk loci after combining results from the current study with the relevant published results. Finally, we conducted GWA analysis of the observed data in the Belgian cohort to see whether there were any novel loci associated with endometriosis risk identified in this sample.
Materials and Methods
Study Participants
The cases (n = 1,077) and controls (n = 900) who were recruited in this study at the Leuven University Hospital, Belgium during 1993–2012 had undergone laparoscopy for sub-fertility with or without pain. Presence of endometriosis in cases was confirmed laparoscopically and histologically based on electronic medical file records. The disease severity in women with endometriosis was prospectively graded according to the rAFS classification system. Endometriosis cases had either minimal (stage I, n = 380), mild (stage II, n = 229), moderate (stage III, n = 174), severe (stage IV, n = 284), or unknown (n = 10) disease. Absence of endometriosis in controls was confirmed laparoscopically. Both cases and controls were Caucasian in origin. All the study participants provided written informed consent, and the study was approved by the Commission of Medical Ethics of the Leuven University Hospital, Belgium and QIMR Berghofer Human Ethics Research Committee, Australia.
DNA Extraction and Genotyping
DNA was purified from EDTA-stabilized whole blood collected for routine molecular diagnostic tests at the Centre for Human Genetics of University Hospitals, Leuven, Belgium. Following the manufacturer's protocol, DNA was purified using Chemagic DNA blood special kit (Chemagen MSM I, PerkinElmer Chemagen Technologies GmbH, Baesweiler, Germany) based on the specific binding of DNA to paramagnetic beads, and Auto Pure LS Puregene chemistry (Qiagen, Venlo, The Netherlands) based on salting-out extraction and a manual salting-out procedure (home-brew). The choice of the extraction method was based on the available amount of blood and on the type of required molecular diagnostic test. DNA concentration was measured using Victor (PerkinElmer, Massachussetts, USA).
Whole genome genotyping of the DNA samples was performed using the Illumina HumanCoreExome 12v1.1 array at the Molecular Epidemiology Laboratory, QIMR Berghofer Medical Research Institute, Brisbane, Australia, following the manufacturer's standard protocol. For a quality control (QC) check, DNA concentrations of majority of the samples were re-measured on the BioTech Powerwave at the QIMR Molecular Epidemiology Laboratory before genotyping. The Illumina HumanCoreExome genotyping arrays are the newer generation of Illumina GWA arrays, which comprised ~250,000 common tag SNPs (‘core’) and ~250,000 predominantly rare coding variants (‘exome’). The exome variants in the Illumina HumanCoreExome were carefully selected based on exome sequencing data in ~12,000 individuals.
Genotype Calling and Quality Control
Genotype data were called using a custom cluster file generated using ~2,000 good quality (<1% missing rate) samples and the GenCall algorithm within Illumina Genome Studio. Data were then further processed by zCall (Goldstein et al., Reference Goldstein, Crenshaw, Carey, Grant, Maguire, Fromer and Neale2012), a rare variant caller, in an attempt to recall missing genotypes. Following manufacturer's guidelines and the protocols developed for the Exome chip data, quality control measures were applied to the Belgian GWA data. Briefly, samples with >1% missing rates, outlying heterozygosity, non-European ancestries (based on 1,000 Genomes Project's European populations), cryptic relatedness (pi-hat > 0.2), and gender discordances were excluded. Similarly, markers with poor separation of three genotype clusters, excess heterozygosity, outlying mean theta and intensity values for heterozygote genotypes, >1% missing rates, the Hardy–Weinberg Equilibrium (HWE) p < 10−6 in controls, and minor allele frequency (MAF) < .05% in either cases or controls were dropped.
Association Analysis
Since rs1096523 in the CDKN2BAS gene at 9p21.3 is monomorphic in populations of European ancestry, we considered the remaining nine SNP loci (rs7521902, rs13394619, rs4141819, rs6542095, rs1519761, rs7739264, rs12700667, rs1537377, and rs10859871) for further analysis. To see whether there were any novel association signals for endometriosis in the Belgian cohort, GWA analysis of the observed genotypes was performed using –assoc (for ‘core’ SNPs) and –fisher (for ‘exome’ variants) commands in Plink for data, including all endometriosis cases (‘All’) and controls. Considering the relatively greater genetic loading of moderate-to-severe (rAFS stage III/IV or ‘Grade_B’) endometriosis compared with mild or minimal (rAFS stage I/II or ‘Grade_A’) disease (Nyholt et al., Reference Nyholt, Low, Anderson, Painter, Uno, Morris and Montgomery2012; Painter et al., Reference Painter, Anderson, Nyholt, Macgregor, Lin, Lee and Zondervan2011; Sapkota et al., Reference Sapkota, Attia, Gordon, Henders, Holliday, Rahmioglu and Nyholt2015a), additional analysis for ‘Grade_B’ endometriosis cases versus controls was also performed. Strengths of association of SNPs with endometriosis are reported in terms of odds ratio (ORs) and confidence intervals (CIs).
Imputation and Meta-Analysis
Of the nine SNPs, only six (rs7521902, rs13394619, rs4141819, rs6542095, rs7739264, and rs12700667) are assayed on Illumina HumanCoreExome 12v1.1 genotyping platform. Therefore, we imputed genotypes in chromosomes containing the nine SNP loci in the Belgian GWA data, using a reference panel of 1,000 Genomes Project (March 2012 release). Imputation was carried out using SHAPEIT (Delaneau et al., Reference Delaneau, Marchini and Zagury2012) and minimac programs (Li et al., Reference Li, Willer, Sanna and Abecasis2009, Reference Li, Willer, Ding, Scheet and Abecasis2010) and following the two-step approach outlined in the online Minimac: 1,000 Genomes Imputation Cookbook (http://genome.sph.umich.edu/wiki/Minimac:_1000_Genomes_Imputation_Cookbook). Quality of the imputed genotypes was assessed by r 2 metric, which estimates the squared correlation between true and imputed genotypes. Poorly imputed SNPs indicated by r 2 < .3 were excluded from downstream analyses. Association analyses of imputed genotype dosage scores of the nine SNP loci were conducted using Plink for ‘All’ and ‘Grade_B’ endometriosis cases separately.
After combining results of imputed dosage scores for the nine SNP loci from Belgian data with the published results of Nyholt et al. (Reference Nyholt, Low, Anderson, Painter, Uno, Morris and Montgomery2012), Adachi et al. (Reference Adachi, Tajima, Quan, Haino, Yoshihara, Masuzaki and Tanaka2010), Albertsen et al. (Reference Albertsen, Chettier, Farrington and Ward2013), and Sapkota et al. (Reference Sapkota, Low, Attia, Gordon, Henders, Holliday and Nyholt2015b), we performed meta-analysis for ‘All’ endometriosis cases and controls. A brief summary of the datasets used in this study is provided in Table 1. Results from Nyholt et al. (Reference Nyholt, Low, Anderson, Painter, Uno, Morris and Montgomery2012) included summary statistics of rs7521902, rs13394619, rs4141819, rs7739264, rs12700667, rs1537377, and rs10859871 obtained from the European (QIMRHCS+OX) and Japanese (BBJ) GWA data (Table 1). Similarly, we included results of rs13394619 and rs6542095 from Adachi et al. (Reference Adachi, Tajima, Quan, Haino, Yoshihara, Masuzaki and Tanaka2010), obtained from the combined analysis of Affymetrix 500K and 6.0 arrays in 696 cases and 825 controls of Japanese ancestry. Furthermore, results from Albertsen et al. (Reference Albertsen, Chettier, Farrington and Ward2013) included summary statistics of rs1519761 in their discovery and replication stages. Finally, we obtained summary results of rs6542095 in the QIMRHCS, OX, and BBJ imputed data from Sapkota et al. (Reference Sapkota, Low, Attia, Gordon, Henders, Holliday and Nyholt2015b).
TABLE 1 Summary of the Datasets Used in the Current Study

QIMRHCS = Queensland Institute of Medical Research and Hunter Community Study; OX = Oxford (UK); BBJ = BioBank of Japan.
Initial meta-analysis was conducted using a fixed-effect (inverse variance-weighted) model implemented in the GWAMA program (Magi & Morris, Reference Magi and Morris2010). Heterogeneity of allelic associations was examined using the Cochran's Q statistic p het < .1 (Cochran, Reference Cochran1954), as well as the I 2 index (Ioannidis et al., Reference Ioannidis, Patsopoulos and Evangelou2007), which indicates the proportion of variance attributable to between-study heterogeneity. Meta-analysis of SNPs associated in fixed-effect model with an evidence of heterogeneity (p < .1) was carried out using the Han–Eskin random-effects model (RE2; Han & Eskin, Reference Han and Eskin2011) implemented in the METASOFT program. In contrast with the conventional random-effects model, the RE2 model increases power under heterogeneity. Furthermore, additional meta-analysis for the nine SNP loci was also performed by restricting to ‘Grade_B’ endometriosis cases (wherever available) versus controls.
Results
Following the QC steps, a total of 998 endometriosis cases and 783 disease-free controls with 316,467 markers remained in the Belgian GWA data for downstream analysis. Of these, 246,071 were ‘core’ SNPs whereas 70,396 were ‘exome’ variants. The GWA analysis of observed genotypes of the 316,467 markers in the Belgian GWA study alone did not produce any genome-wide significant hits in either ‘All’ or ‘Grade_B’ analysis, with few suggestive (p < 1 × 10−5) associations (data not shown). All nine SNPs were accurately imputed with r 2 > .95. We also compared imputed genotypes (dosage scores) of six SNP loci (rs7521902, rs13394619, rs4141819, rs6542095, rs7739264, and rs12700667) for endometriosis with the observed true genotypes available in the Belgian data. Genotype concordances (as measured by the Pearson's correlation coefficient) between two sets of genotypes for the six SNPs were >0.99 (p < 2.2 × 10−16).
Association analysis of the dosage scores of the nine implicated SNP loci in the Belgian data provided further insights into the associations of these SNPs with endometriosis. Risk alleles and their frequencies of all nine SNPs were similar to the ones reported in the original studies (Table 2; Albertsen et al., Reference Albertsen, Chettier, Farrington and Ward2013; Nyholt et al., Reference Nyholt, Low, Anderson, Painter, Uno, Morris and Montgomery2012; Painter et al., Reference Painter, Anderson, Nyholt, Macgregor, Lin, Lee and Zondervan2011; Sapkota et al., Reference Sapkota, Low, Attia, Gordon, Henders, Holliday and Nyholt2015b) and their associations were stronger with ‘Grade_B’ than ‘All’ endometriosis. Furthermore, effect directions of seven out of nine tested SNPs in either ‘All’ or ‘Grade_B’ endometriosis were in line with the published results. Three SNPs showed statistically significant association with endometriosis in either ‘All’ or ‘Grade_B’ disease at a nominal p < .05. SNP rs7521902 showed borderline marginal association (OR = 1.13; p = .12) with ‘All’ endometriosis. As expected, its association was stronger and statistically significant (OR = 1.30; p = .007) with ‘Grade_B’ cases. A statistically significant association (OR = 1.14; p = .045) for rs13394619 was also observed for ‘All’ endometriosis; however, the signal was slightly weaker (OR = 1.13; p = .164) in ‘Grade_B’ cases. A borderline association (OR = 1.14; p = .06) with ‘All’ endometriosis was observed for rs6542095, which was stronger and significant (OR = 1.26; p = .01) in ‘Grade_B’ cases.
TABLE 2 Summary Results of the Nine Known SNP Loci for Endometriosis in the Current Study
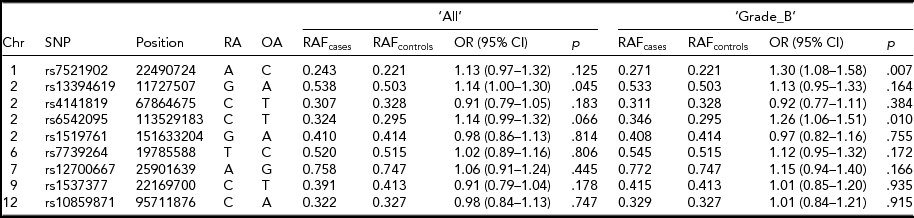
Chr = chromosome; Position = chromosomal position (bp) based on Human Build 37 (GRCh37/hg19); RA = risk allele from original study; OA = other allele; OR = odds ratio; CI = confidence interval.
Meta-analysis, including imputed data from Belgian cohort and the published results, provide insights into SNP loci associated with endometriosis. Six SNP loci showed associations with either ‘All’ or ‘Grade_B’ endometriosis at genome-wide significance level (p < 5 × 10−8) and with similar directions of effect across all studies included in the analysis (Table 3). Of these, three SNPs were associated with both ‘All’ and ‘Grade_B’ endometriosis, with a genome-wide significant evidence in the fixed-effect meta-analysis. These included: SNP rs7521902 near WNT4 (‘All’, OR = 1.17; 95% CI = 1.11–1.23; p = 3.63 × 10−8; ‘Grade_B’, OR = 1.25; 95% CI = 1.17–1.34; p = 1.72 × 10−10), rs13394619 in GREB1 (‘All’, OR = 1.15; 95% CI = 1.10–1.20; p = 9.13 × 10−9; ‘Grade_B’, OR = 1.17; 95% CI = 1.11–1.24; p = 3.02 × 10−8), and rs12700667 at 7p15.2 (‘All’, OR = 1.19; 95% CI = 1.13–1.26; p = 7.10 × 10−10; ‘Grade_B’, OR = 1.29; 95% CI = 1.20–1.39; p = 1.47 × 10−11). The IL1A SNP (rs6542095) was genome-wide significantly associated with only ‘Grade_B’ (OR = 1.22; 95% CI = 1.14–1.30; p = 1.00 × 10−9) endometriosis in fixed-effect meta-analysis, but after appropriate modeling for between-study heterogeneity in ‘All’ endometriosis (p het = .007) in the RE2 model, the association reached genome-wide significance (p = 3.35 × 10−8). Statistical significance of association of rs6542095 with ‘Grade_B’ endometriosis also became stronger (p = 4.90 × 10−10) in the RE2 model after accounting for between-study heterogeneity (p het = .01). A strong association between rs7739264 near ID4 and ‘All’ endometriosis (p = 1.93 × 10−7) was observed, and the signal was further enriched in ‘Grade_B’ endometriosis (OR = 1.20; 95% CI = 1.12–1.27; p = 1.98 × 10−8), achieving a genome-wide significance. Similarly, near genome-wide significant evidence for association between rs1537377 near CDKN2B-AS1 and ‘Grade_B’ endometriosis (OR = 1.19; 95% CI = 1.12–1.27; p = 9.27 × 10−8) was observed, which was genome-wide significant (p = 4.80 × 10−8) after modeling for borderline between-study heterogeneity (p het = .1). However, the effect of rs1537377 in ‘All’ endometriosis was in opposite direction of the published results (Table 2).
TABLE 3 Meta-Analysis for the Nine Known Endometriosis SNP Loci After Combining Summary Statistics From Current Study With the Published Results
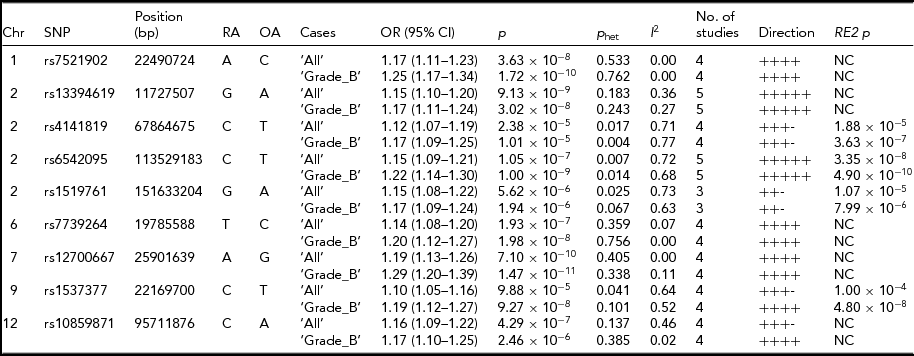
Chr = chromosome; Position = chromosomal position (bp) based on Human Build 37 (GRCh37/hg19); RA = risk allele; OA = other allele; OR = odds ratio; CI = confidence interval; P het = Cochran's Q between-study heterogeneity test p value; I 2, percentage of variance attributable to between-study heterogeneity; RE2 = Han Eskin's random effects model meta-analysis; NC = not calculated.
While the remaining three SNPs (rs4141819, rs1519761, and rs10859871) did not produce genome-wide significant evidence for association with either ‘All’ or ‘Grade_B’ endometriosis in fixed-effect meta-analysis, they still showed strong associations with the disease (p < 2.38 × 10−5). Nonetheless, the effects of rs4141819 and rs1519761 in both ‘All’ and ‘Grade_B’ endometriosis, and that of rs10859871 in ‘All’ endometriosis were in opposite directions of the published results (Table 2). SNP rs4141819 showed between-study heterogeneity (p het < .01) in both ‘All’ and ‘Grade_B’ endometriosis, and after accounting for this heterogeneity in the RE2 model, association of rs4141819 with ‘Grade_B’ disease became stronger with near genome-wide significant evidence (p = 3.63 × 10−7). Significant between-study heterogeneity for rs1519761 was also observed in both ‘All’ and ‘Grade_B’ endometriosis, but its association with the disease (‘All’, p = 5.62 × 10−6; ‘Grade_B’, p = 1.94 × 10−6) slightly diluted in the RE2 model (‘All’, p = 1.07 × 10−5; ‘Grade_B’, p = 7.99 × 10−6). A near genome-wide significant association between rs10859871 near VEZT and ‘All’ endometriosis (OR = 1.16; 95% CI = 1.09–1.22; p = 4.29 × 10−7) was observed, with slightly larger effect size (OR = 1.17; 95% CI = 1.10–1.25) in ‘Grade_B’ disease, although statistical significance of the signal was weaker (p = 2.46 × 10−6).
Discussion
Endometriosis is a complex disease and studies have shown that genetic risk factors substantially contribute to the risk of endometriosis. Genetic studies, especially the GWA studies for endometriosis, have identified 10 SNP loci, of which nine are polymorphic in the populations of European origin (Albertsen et al., Reference Albertsen, Chettier, Farrington and Ward2013; Nyholt et al., Reference Nyholt, Low, Anderson, Painter, Uno, Morris and Montgomery2012; Painter et al., Reference Painter, Anderson, Nyholt, Macgregor, Lin, Lee and Zondervan2011; Sapkota et al., Reference Sapkota, Low, Attia, Gordon, Henders, Holliday and Nyholt2015b). While much larger and well-powered GWA studies are needed to identify additional genetic risk factors involved in the risk of endometriosis, replication studies are crucial to provide credibility that the initial genotype–phenotype associations are valid. Repeated observation of such associations in independent populations of similar ethnicity adds evidence that the associations are not due to chance alone. The previous three replication studies for endometriosis have investigated only a handful of the 10 implicated SNP loci to date. Here we report the most comprehensive replication study performed to date, in which we examine all nine implicated SNP risk loci for endometriosis that are polymorphic in populations of European ancestry, by utilizing GWA data in uniquely characterized 998 endometriosis cases and 783 controls from Belgium.
The risk alleles and their frequencies for all the nine SNPs in the Belgian replication cohort were comparable with the original studies (Table 2; Albertsen et al., Reference Albertsen, Chettier, Farrington and Ward2013; Nyholt et al., Reference Nyholt, Low, Anderson, Painter, Uno, Morris and Montgomery2012; Painter et al., Reference Painter, Anderson, Nyholt, Macgregor, Lin, Lee and Zondervan2011; Sapkota et al., Reference Sapkota, Low, Attia, Gordon, Henders, Holliday and Nyholt2015b). Moreover, direction of effects for seven of the nine SNPs for either ‘All’ or ‘Grade_B’ endometriosis was also consistent with the published results. Among these, we could successfully replicate associations of three SNPs (rs7521902, rs13394619, and rs6542095) with either ‘All’ or ‘Grade_B’ endometriosis at nominal p < .05, which is more often than by chance alone (p = .008; one-sided binomial test). Significant association of rs6542095 at the IL1A locus with ‘All’ (p = .066) and ‘Grade_B’ (p = .01) endometriosis is noteworthy as this is the first successful replication in an independent population, providing further supporting evidence for a potential link between inflammation and endometriosis pathogenesis. More importantly, all the SNPs showed larger effect sizes with ‘Grade_B’ than ‘All’ endometriosis — an observation consistent with the previous reports supporting greater genetic loading in moderate-to-severe disease (Nyholt et al., Reference Nyholt, Low, Anderson, Painter, Uno, Morris and Montgomery2012; Painter et al., Reference Painter, Anderson, Nyholt, Macgregor, Lin, Lee and Zondervan2011; Sapkota et al., Reference Sapkota, Attia, Gordon, Henders, Holliday, Rahmioglu and Nyholt2015a).
Our meta-analysis, including results from the current replication study and the published results, produced genome-wide significant evidence for six (rs7521902 near WNT4, rs13394619 in GREB1, rs6542095 in IL1A, rs7739264 near ID4, rs12700667 at 7p15.2, and rs1537377 near CDKN2B-AS1) of the nine implicated SNPs in either ‘All’ or ‘Grade_B’ endometriosis, after accounting for between-study heterogeneity using the RE2 model, wherever appropriate (Table 3). With the exception of rs1519761 at 2q23.3 reported by Albertsen et al. (Reference Albertsen, Chettier, Farrington and Ward2013), the other two SNP loci (rs4141819 at 2p14 and rs10859871 near VEZT) also showed near genome-wide significance for ‘Grade_B’ endometriosis in the RE2 model (p = 3.63 × 10−7) and for ‘All’ endometriosis in the fixed-effect model (p = 4.29 × 10−7). The association signal for rs1519761 was the weakest (‘All’, p = 5.62 × 10−6; ‘Grade_B’, p = 1.94 × 10−6) among the nine risk loci, and the signal was slightly diluted after accounting for observed between-study heterogeneity in the RE2 model (‘All’, p = 1.07 × 10−5; ‘Grade_B’, p = 7.99 × 10−6). Association signal at this locus was also not replicated in a recent meta-analysis for endometriosis (Rahmioglu et al., Reference Rahmioglu, Nyholt, Morris, Missmer, Montgomery and Zondervan2014), suggesting that further investigation is required to confirm a role for this locus in the risk of endometriosis.
In our multi-ethnic GWA meta-analysis that strongly associated seven risk loci with endometriosis, we found stronger associations at six loci (rs56318008 at 1p36.12, rs77294520 at 2p25.1, rs2861694 at 2p14, rs6901079 at 6p22.3, rs7041895 at 9p21.3, and rs11107968 at 12q22) when we imputed genotypes in the region 2,500 kb upstream and downstream of the most significant genotyped SNP using the full reference panel from the 1,000 Genomes Project Interim Phase 1 Haplotypes (2010–2011 data freeze). For the risk loci at 7p15.2, the genotyped SNP rs12700667 was the best signal. For the remaining six loci with stronger association signals (‘best’ SNPs) post-imputation than the genotyped SNP, we assessed for their replication in the Belgian cohort (Table 4). All six SNPs were accurately imputed in the current study with r 2 > .85. Association results were consistent with that of the original genotyped SNPs, as shown in Table 2, in particular for SNPs rs56318008 at 1p36 and rs77294520 at 2p25.1, which showed nominally significant associations with ‘All’ (p < .098) and ‘Grade_B’ (p < .051), providing further supporting evidence for implication of these risk loci in endometriosis.
TABLE 4 Association Statistics for the ‘Best’ SNPs at the Six Genome-Wide Significant Loci for Endometriosis Reported in Nyholt et al. (Reference Nyholt, Low, Anderson, Painter, Uno, Morris and Montgomery2012)
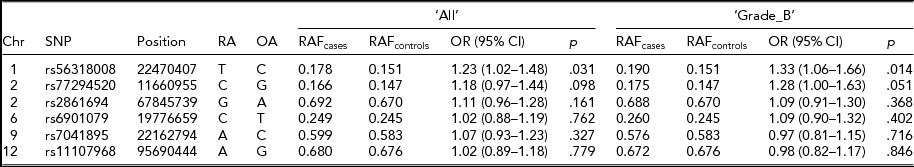
Chr = chromosome; Position = chromosomal position (bp) based on Human Build 37 (GRCh37/hg19); RA = risk allele; OA = other allele; OR = odds ratio; CI = confidence interval.
As a first step to help identify causal variants at nine SNP loci, we interrogated the ExomeChip data for putatively functional coding variants within genes harboring or closest to GWA SNPs. For the GREB1 locus, we found another coding variant, rs10929757, showing nominally significant association (p = .015) with endometriosis. The effect size of rs10929757 was similar (OR = 1.18) to the GWA SNP rs13394619, although they are poorly correlated (r 2 = .25). Similarly, we also observed nominally significant association (p < .018) for two coding variants (rs2383207 and rs4977574) in CDKN2B-AS1 — the closest gene to the GWA SNP rs1537377 at 9p21.3. In spite of lack of correlation (r 2 = .011 and .005, respectively) with rs1537377, the effect sizes for both variants were similar (ORs = 1.17 and 1.19 respectively). While these data may suggest independent association signals at GREB1 and 9p21.3 loci, the three coding variants need to be further investigated in a larger sample size for a more conclusive interpretation. We did not observe evidence of association for other coding variants at nominal p < .05, even though the effect sizes for some were comparable with GWA SNPs at each risk loci (data not shown). We did not detect any rare coding variants at GWA loci despite the ExomeChip data. This may be due, in part, to reduced power in the Belgian sample to detect such rare variants. Assuming a disease prevalence of 8%, our sample size only had 45% power to detect alleles of frequency .20 contributing to genotype relative risk of 1.15 (Purcell et al., Reference Purcell, Cherny and Sham2003). As such, larger ExomeChip studies may be required to adequately investigate potential role of coding/rare variants in the risk of endometriosis and other complex traits. We cannot rule out the possibility of other types of rare functional variants at these loci, which are not adequately captured by either ExomeChip or current imputation methods, contributing to increased risk of endometriosis. These issues may be addressed by the future studies utilizing larger sample sizes, coupled with re-sequencing and further fine-mapping required to identify causal variants within the implicated GWA loci. Furthermore, controls used in this study were clinic-based endometriosis-free individuals who presented with symptoms of sub-fertility. As such, they may have different allele frequencies as compared with ‘population-based’ controls used in most GWA studies, and hence may partly explain the opposite direction of effect sizes observed for some SNPs. However, this needs to be investigated further using a larger sample size with population-based controls, and therefore caution should be used interpreting these results.
Overall, results from the current replication study provide further supporting evidence for associations of the implicated SNP loci with endometriosis. Meta-analysis for these loci after including additional published results produced genome-wide significant evidence for six loci, with similar magnitudes and directions of effect across studies, and hence provided further evidence against any possibility of inflated genetic effects due to the ‘winner's curse’ bias in the original study. More importantly, all the nine SNPs showed larger effect sizes with stage III/IV endometriosis than all cases, corroborating our previous observation for greater genetic loading in moderate-to-severe endometriosis.
Acknowledgments
We would like to thank all the participants in the endometriosis studies who were included in this analysis. We also thank many hospital directors and staff, gynecologists, general practitioners, and pathology services who provided assistance with confirmation of diagnoses. Dale R. Nyholt was supported by an NHMRC Fellowship (613674) and ARC Future Fellowship (FT0991022) schemes, and Grant W. Montgomery was supported by the NHMRC Fellowships Scheme (339446, 619667).






