The timing of the sleep–wake cycle is regulated by the body's endogenous circadian rhythm system as well as sleep homeostatic processes (Panda et al., Reference Panda, Hogenesch and Kay2002). Although environmental cues entrain phase and length of sleep, there is substantial variability in the timing of the biological clock in individuals. This ‘temporal phenotype of an organism’ is now commonly referred to as a chronotype (Ehret, Reference Ehret1974). An element of chronotype, diurnal preference, is believed to reflect endogenous circadian rhythms. People colloquially referred to as a morning-person (or lark) are more alert in the morning and fall asleep easily at night, and an evening-person (or owl) is more alert in the evening and can go to bed and wake later.
Natural variation in circadian rhythmicity can contribute to disparities in fitness within species; variations in circadian period length may influence a variety of physiological and behavioral characteristics. Lifespan decreases as the magnitude of circadian period length deviation from the 24-hour cycle increases in rodents, and primate species (Kerkhof, Reference Kerkhof1985; Partonen, Reference Partonen2013; Wyse et al., Reference Wyse, Coogan, Selman, Hazlerigg and Speakman2010). Evening preference is associated with several psychosocial and psychopathological issues (Fabbian et al., Reference Fabbian, Zucchi, De Giorgi, Tiseo, Boari, Salmi and Manfredini2016; Fares et al., Reference Fares, Hermens, Naismith, White, Hickie and Robillard2015; Haregu et al., Reference Haregu, Gelaye, Pensuksan, Lohsoonthorn, Lertmaharit, Rattananupong and Williams2015; Merikanto et al., Reference Merikanto, Suvisaari, Lahti and Partonen2016; Rose et al., Reference Rose, Gelaye, Sanchez, Castaneda, Sanchez, Yanez and Williams2015; Voinescu et al., Reference Voinescu, Szentagotai and David2012), including depression (Abe et al., Reference Abe, Inoue, Komada, Nakamura, Asaoka, Kanno and Takahashi2011; Antypa et al., Reference Antypa, Vogelzangs, Meesters, Schoevers and Penninx2016; Chan et al., Reference Chan, Lam, Li, Yu, Chan, Zhang and Wing2014; Chelminski et al., Reference Chelminski, Ferraro, Petros and Plaud1999; Drennan et al., Reference Drennan, Klauber, Kripke and Goyette1991; Gaspar-Barba et al., Reference Gaspar-Barba, Calati, Cruz-Fuentes, Ontiveros-Uribe, Natale, De Ronchi and Serretti2009; Hidalgo et al., Reference Hidalgo, Caumo, Posser, Coccaro, Camozzato and Chaves2009; Kitamura et al., Reference Kitamura, Hida, Watanabe, Enomoto, Aritake-Okada, Moriguchi and Mishima2010; Levandovski et al., Reference Levandovski, Dantas, Fernandes, Caumo, Torres, Roenneberg and Allebrandt2011; Merikanto et al., Reference Merikanto, Lahti, Kronholm, Peltonen, Laatikainen, Vartiainen and Partonen2013; Reference Merikanto, Kronholm, Peltonen, Laatikainen, Vartiainen and Partonen2015; Muller et al., Reference Muller, Cabanel, Olschinski, Jochim and Kundermann2015; Reid et al., Reference Reid, Jaksa, Eisengart, Baron, Lu, Kane and Zee2012), bipolar disorder (BD; Ahn et al., Reference Ahn, Chang, Joo, Kim, Lee and Kim2008; Giglio et al., Reference Giglio, Magalhaes, Andersen, Walz, Jakobson and Kapczinski2010; Jeong Jeong et al., Reference Jeong Jeong, Moon, Min Park, Dae Lee, Min Lee, Choi and In Chung2015; Mansour et al., Reference Mansour, Wood, Chowdari, Dayal, Thase, Kupfer and Nimgaonkar2005; Melo et al., Reference Melo, Abreu, Linhares Neto, de Bruin and de Bruin2016; Seleem et al., Reference Seleem, Merranko, Goldstein, Goldstein, Axelson, Brent and Birmaher2015; Wood et al., Reference Wood, Birmaher, Axelson, Ehmann, Kalas, Monk and Nimgainkar2009), and substance use (Adan, Reference Adan1994; Barclay et al., Reference Barclay, Eley, Parsons, Willis and Gregory2013; Broms et al., Reference Broms, Kaprio, Hublin, Partinen, Madden and Koskenvuo2011; Hasler et al., Reference Hasler, Sitnick, Shaw and Forbes2013; Kervran et al., Reference Kervran, Fatseas, Serre, Taillard, Beltran, Leboucher and Auriacombe2015; Lemoine et al., Reference Lemoine, Zawieja and Ohayon2013; Reid et al., Reference Reid, Jaksa, Eisengart, Baron, Lu, Kane and Zee2012; Sivertsen et al., Reference Sivertsen, Skogen, Jakobsen and Hysing2015).
Genetic epidemiologic estimates of the additive heritability of diurnal preferences range from 21% to 54% (Barclay et al., Reference Barclay, Eley, Buysse, Archer and Gregory2010; Evans et al., Reference Evans, Snitker, Wu, Mody, Njajou, Perlis and Hsueh2011; Heath et al., Reference Heath, Kendler, Eaves and Martin1990; Hur et al., Reference Hur, Bouchard and Lykken1998; Klei et al., Reference Klei, Reitz, Miller, Wood, Maendel, Gross and Nimgaonkar2005; Koskenvuo et al., Reference Koskenvuo, Hublin, Partinen, Heikkila and Kaprio2007; Toomey et al., Reference Toomey, Panizzon, Kremen, Franz and Lyons2015; Vink et al., Reference Vink, Groot, Kerkhof and Boomsma2001; von Schantz et al., Reference von Schantz, Taporoski, Horimoto, Duarte, Vallada, Krieger and Pereira2015; Watson et al., Reference Watson, Buchwald and Harden2013). The results of several genome-wide association studies (GWAS) on sleep duration, sleep latency, sleep depth, sleep quality, insomnia, daytime sleepiness, sleep apnea, and actigraphic measurements are available (Allebrandt et al., Reference Allebrandt, Amin, Muller-Myhsok, Esko, Teder-Laving, Azevedo and Roenneberg2013; Amin et al., Reference Amin, Allebrandt, van der Spek, Muller-Myhsok, Hek, Teder-Laving and van Duijn2016; Byrne et al., Reference Byrne, Gehrman, Medland, Nyholt, Heath, Madden and Chronogen2013; Cade, et al., Reference Cade, Chen, Stilp, Gleason, Sofer, Ancoli-Israel and Redline2016; Cade et al., Reference Cade, Chen, Stilp, Gleason, Sofer, Ancoli-Israel and Redline2016; Gottlieb et al., Reference Gottlieb, O'Connor and Wilk2007; Reference Gottlieb, Hek, Chen, Watson, Eiriksdottir, Byrne and Tiemeier2015; Lane et al., Reference Lane, Liang, Vlasac, Anderson, Bechtold, Bowden and Saxena2017; Marinelli et al., Reference Marinelli, Pappa, Bustamante, Bonilla, Suarez, Tiesler and Sunyer2016; Ollila et al., Reference Ollila, Kettunen, Pietilainen, Aho, Silander, Kronholm and Paunio2014; Scheinfeldt et al., Reference Scheinfeldt, Gharani, Kasper, Schmidlen, Gordon, Jarvis and Christman2015; Spada et al., Reference Spada, Scholz, Kirsten, Hensch, Horn, Jawinski and Sander2016), yet many of these phenotypes are independent of chronotype (Jones et al., Reference Jones, Tyrrell, Wood, Beaumont, Ruth, Tuke and Weedon2016; Lane et al., Reference Lane, Liang, Vlasac, Anderson, Bechtold, Bowden and Saxena2017; Lehnkering & Siegmund, Reference Lehnkering and Siegmund2007). Two early GWAS identified several single nucleotide polymorphisms (SNPs) associated with the chronotype measure bedtime. Only an intronic variant in erythrocyte membrane protein band 4.1, EPB41, was nominally associated in both studies. The EPB41 linked variant is not significantly associated after correction for multiple testing (Byrne et al., Reference Byrne, Gehrman, Medland, Nyholt, Heath, Madden and Chronogen2013; Gottlieb et al., Reference Gottlieb, O'Connor and Wilk2007). Many loci associated with chronotype have been detected and replicated by three large-scale GWAS. Hu and colleagues identified 15 genome-wide significant loci using 89,283 subjects from the 23andMe cohort (Hu et al., Reference Hu, Shmygelska, Tran, Eriksson, Tung and Hinds2016). Eight of the 15 loci were subsequently replicated in a sample of 100,420 subjects from the UK Biobank (Lane et al., Reference Lane, Vlasac, Anderson, Kyle, Dixon, Bechtold and Saxena2016). Among these loci, the linked genes for regulator of G-protein signaling 16 (RGS16), vasoactive intestinal peptide (VIP), aph-1 homolog A, gamma-secretase subunit (APH1A), F-box and leucine rich repeat protein 13 (FBXL13), and the period genes PER2 and PER3 have plausible roles sleep biology. Jones et al. (Reference Jones, Tyrrell, Wood, Beaumont, Ruth, Tuke and Weedon2016) used a larger cohort from the UK Biobank (N = 128,266) in their GWAS. This work replicated these findings and confirmed associations detected with the 23andMe cohort (linked to adenylate kinase 5 (AK5) and hypocretin receptor 2 (HCRTR2; Hu et al., Reference Hu, Shmygelska, Tran, Eriksson, Tung and Hinds2016)), and identified seven additional loci that were genome-wide significant in both the UK Biobank cohort and meta-analysis of the UK Biobank and 23andME cohorts (linked to exonuclease 3’–5’ domain containing 3 (EXD3), 5-hydroxytryptamine receptor 6 (HTR6), FK506 binding protein 1B (FKBP1B), calbindin 1 (CALB1), PATJ crumbs cell polarity complex component (INADL), and acylphosphatase 2 (ACYP2; Jones et al., Reference Jones, Tyrrell, Wood, Beaumont, Ruth, Tuke and Weedon2016)). While these studies have shed light onto the molecular basis of diurnal preference in human populations, they have solely looked at populations of European ancestry. Several GWAS on sleep phenotypes have included subjects of diverse ethnic background (Cade et al., Reference Cade, Chen, Stilp, Gleason, Sofer, Ancoli-Israel and Redline2016; Scheinfeldt et al., Reference Scheinfeldt, Gharani, Kasper, Schmidlen, Gordon, Jarvis and Christman2015), including a solely Hispanic sample (Cade et al., Reference Cade, Chen, Stilp, Gleason, Sofer, Ancoli-Israel and Redline2016), yet these studies did not assess chronotype.
Over the last decade there has been a growing body of the literature that has explored the potential overlap between health disparities research and sleep medicine (Jean-Louis & Grandner, Reference Jean-Louis and Grandner2016). A number of epidemiological studies have reported poorer quality sleep and a higher prevalence of short and/or long sleep in non-White adults, particularly from lower socioeconomic status groups, when compared to White adults (Carnethon et al., Reference Carnethon, De Chavez, Zee, Kim, Liu, Goldberger and Knutson2016; Hale & Do, Reference Hale and Do2007; Krueger & Friedman, Reference Krueger and Friedman2009; Stamatakis et al., Reference Stamatakis, Kaplan and Roberts2007). Little is known concerning genetic factors that might influence sleep in different ethnic groups.
The purpose of this study was to investigate genetic influences on chronotype in two admixed populations: a young adult sample of Hispanics who predominately identify as Mexican American (MA), and a family-based sample of American Indians (AIs). Recent work from our laboratory has identified specific demographic, clinical, and cultural factors associated with sleep characteristics in these samples (Ehlers et al., Reference Ehlers, Gilder, Criado and Caetano2010; Reference Ehlers, Wills, Lau and Gilder2017). Briefly, in the AI sample, higher degrees of AI heritage, but not cultural identification, being over the age of 30, and having a high school diploma were all predictive of short sleep duration (<6 hours). The global score on the Pittsburgh Sleep Quality Index (PSQI; Buysse et al., Reference Buysse, Reynolds, Monk, Berman and Kupfer1989) was significantly higher in those participants with a lifetime diagnosis of substance use disorders, anxiety disorders, and affective disorders. Alcohol use disorders and affective disorders were significant predictors of sleep latency, whereas anxiety and affective disorders were correlated with waking more often in the night/early morning. Nicotine dependence was associated with having trouble breathing and alcohol use disorders and anxiety disorders with bad dreams (Ehlers et al., Reference Ehlers, Wills, Lau and Gilder2017). In the MA sample, lifetime diagnoses of alcohol-use disorders, family history of alcohol dependence, acculturation stress, and lifetime diagnoses of major depressive disorder were all correlated with significantly poorer quality sleep as indexed by the global score on the PSQI. Regression analyses also revealed that gender was correlated with habitual bedtime, whereas drug dependence (cannabis, stimulants, and/or opiates) was significantly correlated with how long it took to fall asleep, major depressive disorder with the number of hours spent sleeping a night, and anxiety disorders and major depressive disorder with waking up in the early morning or middle of the night (Ehlers et al., Reference Ehlers, Gilder, Criado and Caetano2010).
Previous GWAS on chronotype have exclusively included subjects of European ancestry; hence, two specific analyses in these understudied populations were undertaken in this study. First, we conducted a GWAS on diurnal preference. In existing studies of Europeans, only common sequence variants with small effect have been identified through GWAS. In the second analysis, polygenic risk score profiling was used to determine whether a larger proportion of variance in diurnal preference could be attributed to variants that have been associated with other complex disorders. The following phenotypes were analyzed for overlapping genetic influences with chronotype: BP and depression, given the robust link between these psychiatric disorders and chronotype (Abe et al., Reference Abe, Inoue, Komada, Nakamura, Asaoka, Kanno and Takahashi2011; Ahn et al., Reference Ahn, Chang, Joo, Kim, Lee and Kim2008; Antypa et al., Reference Antypa, Vogelzangs, Meesters, Schoevers and Penninx2016; Chan et al., Reference Chan, Lam, Li, Yu, Chan, Zhang and Wing2014; Chelminski et al., Reference Chelminski, Ferraro, Petros and Plaud1999; Drennan et al., Reference Drennan, Klauber, Kripke and Goyette1991; Gaspar-Barba et al., Reference Gaspar-Barba, Calati, Cruz-Fuentes, Ontiveros-Uribe, Natale, De Ronchi and Serretti2009; Giglio et al., Reference Giglio, Magalhaes, Andersen, Walz, Jakobson and Kapczinski2010; Hidalgo et al., Reference Hidalgo, Caumo, Posser, Coccaro, Camozzato and Chaves2009; Jeong Jeong et al., Reference Jeong Jeong, Moon, Min Park, Dae Lee, Min Lee, Choi and In Chung2015; Kitamura et al., Reference Kitamura, Hida, Watanabe, Enomoto, Aritake-Okada, Moriguchi and Mishima2010; Levandovski et al., Reference Levandovski, Dantas, Fernandes, Caumo, Torres, Roenneberg and Allebrandt2011; Mansour et al., Reference Mansour, Wood, Chowdari, Dayal, Thase, Kupfer and Nimgaonkar2005; Melo et al., Reference Melo, Abreu, Linhares Neto, de Bruin and de Bruin2016; Merikanto et al., Reference Merikanto, Lahti, Kronholm, Peltonen, Laatikainen, Vartiainen and Partonen2013; Reference Merikanto, Kronholm, Peltonen, Laatikainen, Vartiainen and Partonen2015; Muller et al., Reference Muller, Cabanel, Olschinski, Jochim and Kundermann2015; Reid et al., Reference Reid, Jaksa, Eisengart, Baron, Lu, Kane and Zee2012; Seleem et al., Reference Seleem, Merranko, Goldstein, Goldstein, Axelson, Brent and Birmaher2015; Wood et al., Reference Wood, Birmaher, Axelson, Ehmann, Kalas, Monk and Nimgainkar2009); and schizophrenia (SCZ) and body mass index (BMI), given previously identified genetic correlations between these phenotypes and chronotype (Lane et al., Reference Lane, Vlasac, Anderson, Kyle, Dixon, Bechtold and Saxena2016).
Materials and Methods
Samples
Six hundred and nineteen young adult MAs were recruited using a commercial mailing list. The sample consists of primarily second-generation MAs, as previously described (Ehlers et al., Reference Ehlers, Gilder, Criado and Caetano2010). Each subject was required to be of MA heritage, between the ages of 18 and 30, residing in the United States legally, and able to read and write in English to be included in the study; subjects who were pregnant, nursing, or currently had a major medical or neurological disorder were excluded. Hispanic heritage was assessed based on the origin (Caribbean or West Indian, Chicano, Cuban, Mexican, Mexican-American, Puerto Rican, South American, other Spanish, or Mexican Indian) of each subject's eight grandparents; 83.8% of the sample self-identified as having 50% or more Mexican heritage alone.
A family-based sample of AI participants recruited from eight geographically continuous reservations on which roughly 3,000 individuals reside comprised the second sample. To be included in the study, each participant had to be of AI heritage, between the ages of 18 and 82, and be mobile enough to be transported from his or her home to The Scripps Research Institute (TSRI), as previously described (Ehlers et al., Reference Ehlers, Wills, Lau and Gilder2017).
The primary phenotype, whether each subject was an ‘owl’ or not, was indexed by the PSQI (Buysse et al., Reference Buysse, Reynolds, Monk, Berman and Kupfer1989), the psychometric properties of which have been described elsewhere (Carpenter & Andrykowski, Reference Carpenter and Andrykowski1998). The PSQI has over 12,000 citations and has been used in many cultures. The PSQI consists of 19 individual items such as usual bedtime, wake-up time, actual hours slept, number of minutes to fall asleep, and nighttime awakenings. If a subject answered after 11 pm when asked the question ‘When [what time] have you usually gone to bed?’ they were considered an owl, based on Dr Buysse's recommendation of what is standard to use for the definition of evening preference on the PSQI. Extraneous phenotypic information on substance use and various affective disorders for these samples was obtained through interviews using the semi-structured assessment for the Genetics of Alcoholism (SSAGA; Bucholz et al., Reference Bucholz, Cadoret, Cloninger, Dinwiddie, Hesselbrock, Nurnberger and Schuckit1994), which has exhibited reliability and validity for diagnosing these disorders (Bucholz et al., Reference Bucholz, Cadoret, Cloninger, Dinwiddie, Hesselbrock, Nurnberger and Schuckit1994; Hesselbrock et al., Reference Hesselbrock, Easton, Bucholz, Schuckit and Hesselbrock1999).
All work was carried out in accordance with the ethical standards of the relevant national and institutional committees on human experimentation and with the Helsinki Declaration of 1975, as revised in 2008. Written consent was obtained for all participants. The Institutional Review Board (IRB) at TSRI approved the protocol for this study and the Indian Health Council, a tribal review group overseeing health issues for the reservations in which recruitment took place, additionally approved the AI study protocol.
Genotyping
DNA was extracted from consenting subjects from blood samples. In the MA cohort, DNA was prepared and genotyped using the Affymetrix Exome1A chip according to the Affymetrix Axiom 2.0 Assay Manual Workflow documentation. As described previously (Norden-Krichmar et al., Reference Norden-Krichmar, Gizer, Wilhelmsen, Schork and Ehlers2014), quality control on the markers was initially performed according to Affymetrix best practices. Additional quality control measures included: removing SNPs out of Hardy–Weinberg equilibrium (HWE, p < 1E-10), as calculated on a set of maximally unrelated subjects using PLINK (Purcell et al., Reference Purcell, Neale, Todd-Brown, Thomas, Ferreira, Bender and Sham2007); excluding SNPs with bad genotype clusters using snpChecker (a Java program with a graphical interface that allows the investigator to visually check and compare the clustering characteristics of each variants and either recluster bad calls or remove variants with poor overall genotype calls accordingly); removing subjects of high hidden relatedness using genome-wide complex trait analysis (GCTA, with the genetic relationship matrix, GRM, cut-off 0.125; Yang et al., Reference Yang, Lee, Goddard and Visscher2011); and pruning out SNPs with low minor allele frequency (MAF <1%). Imputation was performed using the Michigan Imputation Server (https://imputationserver.sph.umich.edu/index.html#!pages/home); the ShapeIT software was used to phase the data and the 1000 Genomes (1000G) Phase 3 v5 American (AMR) population as a reference panel. Post-imputation quality control steps were performed as follows: removing duplicates, retaining variants with very high imputation accuracy (Rsq ≥ 0.9), removing variants with low MAF (<1%) and variants out of HWE (p < .001); 301,019 variants remained for analysis.
DNA from the AI cohort was also extracted from blood samples. DNA sequencing was done using Illumina low-coverage whole-genome sequencing (WGS) and samples were additionally genotyped using an Affymetrix Exome1A chip, as previously described (Bizon et al., Reference Bizon, Spiegel, Chasse, Gizer, Li, Malc and Wilhelmsen2014). Paired-end sequencing was performed using the HiSeq2000 sequencers. For roughly 80% of samples, the sequencing coverage was approximately evenly distributed at 3–12x. WGS reads were aligned using BWA (Li & Durbin, Reference Li and Durbin2009) and realigned around indels using GATK (DePristo et al., Reference DePristo, Banks, Poplin, Garimella, Maguire, Hartl and Daly2011). Variants were called using two pipelines for comparison: the GATK Unified Genotyper, following best practices for low-coverage samples (Van der Auwera et al., Reference Van der Auwera, Carneiro, Hartl, Poplin, Del Angel, Levy-Moonshine and DePristo2013); and the linkage disequilibrium (LD) aware variant caller Thunder (Li et al., Reference Li, Sidore, Kang, Boehnke and Abecasis2011). Variant calls from Thunder were used in these analyses. The sequencing calls and exome genotypes were in good agreement (Bizon et al., Reference Bizon, Spiegel, Chasse, Gizer, Li, Malc and Wilhelmsen2014). Variant calls for the 301,019 variants imputed in the MA sample were extracted from the AI sequence calls; 269,453 total variants were selected based on base pair location. Quality control steps for this subset of variants included removing variants with low MAF (<1%) and variants out of HWE (p < .001, using a set of maximally unrelated subjects).
The MA and AI data were merged using PLINK (Purcell et al., Reference Purcell, Neale, Todd-Brown, Thomas, Ferreira, Bender and Sham2007), resulting in 258,423 markers, and cleaned as previously: removing variants with low MAF (<1%), and removing variants out of HWE (p < .001, using a set of maximally unrelated subjects). The final number of variants for analysis was 258,411. Lastly, GCTA (Yang et al., Reference Yang, Lee, Goddard and Visscher2011) was used to generate a GRM and 20 principal components (PCs) after the genotype data were pruned for LD.
Genome-Wide Association Analysis
A mixed linear model association (MLMA) (Yang et al., Reference Yang, Zaitlen, Goddard, Visscher and Price2014) was run using GCTA (Yang et al., Reference Yang, Lee, Goddard and Visscher2011). This model tests the additive effect of each SNP for association with the phenotype, given the relationship matrix between subjects (i.e., the GRM calculated in GCTA). Gender, study, age, and 20 PCs were included as covariates. Allele frequencies were calculated within GCTA. Manhattan and quantile–quantile (QQ) plots were generated in R (R Development Core Team, 2012) using the Manhattan package (Turner, Reference Turner2014). The genomic inflation factor (lambda, λ) was also calculated in R using the commands outlined in http://genometoolbox.blogspot.com/2014/08/how-to-calculate-genomic-inflation.html. Locus zoom plots were generated with the LocusZoom software (http://locuszoom.sph.umich.edu/locuszoom/; Pruim et al., Reference Pruim, Welch, Sanna, Teslovich, Chines, Gliedt and Willer2010) using the 1000G AMR population as a reference population and the hg19 build. Variant annotation was performed with ANNOVAR (Wang et al., Reference Wang, Li and Hakonarson2010). Power calculations were executed using the case-control variance components module for discrete traits based upon a formula derived in (Sham et al., Reference Sham, Cherny, Purcell and Hewitt2000), as implemented in the genetic power calculator (Purcell et al., Reference Purcell, Cherny and Sham2003).
Because much fewer variants were tested than in a standard GWAS (which typically includes roughly a million SNPs), the standard Bonferroni correction threshold of p < 5E-08 would be too stringent. Thus, suggestive and genome-wide significance thresholds were calculated based on the effective number of independent tests using the Genetic Type 1 Error Calculator (GEC) software (Li et al., Reference Li, Yeung, Cherny and Sham2012). This software uses LD information from the sample to calculate the number of independent SNPs being tested and calculates suggestive and genome-wide significance thresholds given the number of independent tests. Suggestive and genome-wide significance thresholds were estimated at p < 1.82E-05 and p < 9.11E-07, respectively.
Genetic Risk Score Profiling
Genetic risk scores (GRSs) were generated using PRSice (Euesden et al., Reference Euesden, Lewis and O'Reilly2015). Briefly, to detect shared genetic etiology between traits, GRSs are calculated on a base phenotype using large-scale GWAS results and used as predictors of a target phenotype in individuals from an independent data set. As LD between variants can inflate GRSs, PRSice prunes the base data for LD based on the genotypic structure of the target data (using PLINK 1.9; Chang et al., Reference Chang, Chow, Tellier, Vattikuti, Purcell and Lee2015). PRSice then performs ‘high-resolution’ scoring in which a number of p-value thresholds are used to construct a GRS at each cut-off. Only variants exceeding the set p-value threshold (PT ) are included in the GRS, and each is weighted by its effect size in the base data. Each GRS, which represents a sum of trait-associated alleles in the base data for each p value cut-off, is calculated in the independent target data for each subject. Although by default these scores are then regressed on the phenotype in the target data, for our purposes all scores were written out in a file (i.e., using the commands ‘no.regression T report.best.score F report.individual.scores T’ in PRSice) and regressed on the phenotype using a generalized linear mixed model fit by maximum likelihood. This model was fit using the pedigreemm package (https://cran.r-project.org/web/packages/pedigreemm/pedigreemm.pdf). All default options were used, with the exception of specifying ‘family = binomial’ (to indicate a binary outcome), and a pedigree to be included in the model. Gender, study, age, and 20 PCs were included as fixed effects, with subject fitted as random effects.
Cross-phenotype GRSs were computed for four phenotypes and used as predictors in the model for the owl phenotype: BP, depressive symptoms (DS), SCZ, and BMI. All of these data, described briefly below, were meta-analyses of large-scale GWAS. Summary statistics on BP (Psychiatric GWAS Consortium Bipolar Disorder Working Group, 2011) were made available through the Psychiatric Genomics Consortium (PGC) download page (https://www.med.unc.edu/pgc/results-and-downloads) and were based on a GWAS of 7,481 cases and 9,250 controls. GWAS results of DS in 161,460 subjects (Okbay et al., Reference Okbay, Baselmans, De Neve, Turley, Nivard, Fontana and Cesarini2016) were downloaded from the Social Science Genetic Association Consortium (http://www.thessgac.org/data). The SCZ data (Schizophrenia Working Group of the Psychiatric Genomics Consortium, 2014) were also downloaded from the PGC and consisted of up to 36,989 cases and 113,075 controls. Finally, multi-ethnic summary statistics on BMI from 339,224 individuals were made available through the Genetic Investigation of ANthropometric Traits (GIANT) consortium (Locke et al., Reference Locke, Kahali, Berndt, Justice, Pers, Day and Speliotes2015). For both the BP and SCZ results, risk score training sets were available in which the summary data had already been pruned for LD. For these analyses PRSice's internal clumping procedures were turned off (i.e., clump.snps F prune.snps F). Default clumping procedures in PRSice were used for the DS and BMI results, using the LD structure of the MA/AI target data. GRSs were calculated between p value thresholds of 0.01 and 0.5 at increments of 0.01. The major histocompatibility complex (MHC) region, owing to its complex LD structure, was removed from calculation of the GRS. The suggested significance threshold for a single-phenotype high-resolution GRS analyses after correction for multiple testing is p = .001 (Euesden et al., Reference Euesden, Lewis and O'Reilly2015).
Results
The analyses included 299 AIs (159 cases and 140 controls) and 535 MAs (371 cases and 164 controls) for a total of 834 subjects (Table 1). Generally, the AIs were older, had higher rates of AUDs, higher BMI, fewer years of education, and had a higher percentage of the sample earning less than $20,000 per year compared to the MA cohort. The decision was made to combine the cohorts due to the small sample size of each.
TABLE 1 Phenotypic Characteristics of the Samples

Note: SD = standard deviation, AUD = Alcohol use disorder (mild, moderate, or severe by DSM-5); BMI = body mass index.
Genome-Wide Association Test
Four variants reached the threshold for suggestive evidence for association with being an owl at p < 1.82E-05 (Figure 1). A genomic inflation factor of λ = 0.989 indicated no inflation of test scores (Supplementary Figure S1). All four suggestively significant SNPs were in KIAA1549 like (KIAA1549L), the minor alleles of which showed a consistently negative effect on being an owl (Table 2), and were in high LD (Figure S2). A list of the top 50 associations is provided in Table S1. Post hoc analyses were performed separately in each cohort for the most highly significant SNP. Results indicated that, adjusting for gender, age, 20 PCs, and the GRM, rs10768009 was associated at p < .05 in the same direction in both individual cohorts in a MLMA (for the minor A allele: SNP effect = −0.120, standard error (SE) = 0.029, p = 2.50E-05 in the MAs; and SNP effect = −0.092, SE = 0.043, p = .030 in the AIs). Alcohol use has a well-characterized link to sleep (reviewed in Chakravorty et al., Reference Chakravorty, Chaudhary and Brower2016). The GWAS was repeated including AUD as a covariate (specifically having any AUD, mild, moderate, or severe, by DSM-5); the results were unchanged.
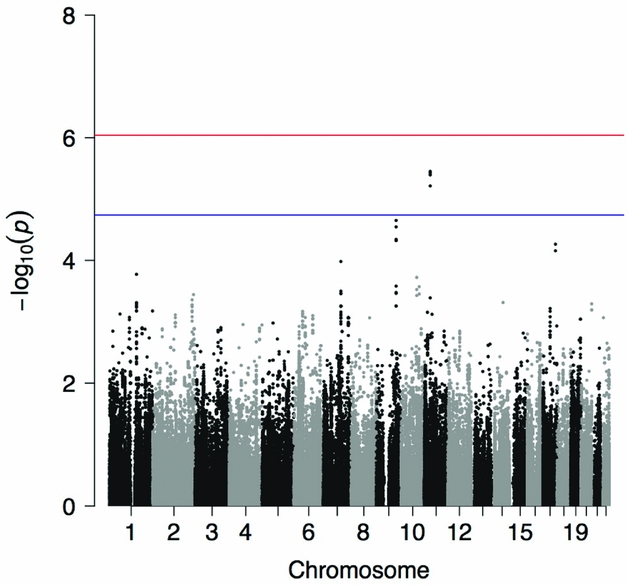
FIGURE 1 Manhattan plot showing the p value for the owl phenotype in the combined American Indian and Mexican American cohorts. Suggestive (blue line) and genome-wide (red line) significance thresholds are p < 1.82E-05 and p < 9.11E-07, respectively, calculated using the Genetic Type 1 Error Calculator (GEC) software.
TABLE 2 Suggestively Significant Hits for the Owl Phenotype in the Combined American Indian and Mexican American Sample

Note: Additional information is available in Supplementary Table S1 and Supplementary Figure S2.
Chr = Chromosome; Freq = Frequency of the effect allele; SE = Standard error; Ref = Reference allele; Alt = Alternate allele; X1000g2014oct_all = Allele frequency of the alternate allele from the 1000 Genomes Project (all populations).
Power calculations for the high-risk C allele were performed with the following criteria: high-risk allele frequency 0.5773; prevalence 0.25; 530 cases; and a 0.574 ratio of controls to cases. For genotype relative risk ratios of 1.5 (heterozygotes) and 1.75 (homozygotes for the risk allele), our study was 12.15% powered to detect an association at p < 1.82E-05. To be 80% powered to detect an effect, 1,432 cases would be needed at p < 1.82E-05 and 1,803 cases would be needed at p < 9.11E-07.
Replication of Previous Associations
Association summary statistics are displayed for SNPs annotated to some of the genes significantly associated with chronotype from previous studies in Table S2: RGS16, AK5, HCRTR2, and FBXL13 (Hu et al., Reference Hu, Shmygelska, Tran, Eriksson, Tung and Hinds2016; Jones et al., Reference Jones, Tyrrell, Wood, Beaumont, Ruth, Tuke and Weedon2016; Lane et al., Reference Lane, Vlasac, Anderson, Kyle, Dixon, Bechtold and Saxena2016). No SNPs were mapped to APH1A. Of the SNPs previously associated with chronotype, only one was directly assessed in the current study: rs1144566 in RGS16 was associated at p < 1.3E-08 in the study by Lane et al. (Reference Lane, Vlasac, Anderson, Kyle, Dixon, Bechtold and Saxena2016) but failed to reach nominal significance in our study with p > .1. In fact, none of the SNPs mapped to these genes were associated with being an owl at p < .05; however, evaluation of LD using SNP Annotation and Proxy Search (SNAP; Johnson et al., Reference Johnson, Handsaker, Pulit, Nizzari, O'Donnell and de Bakker2008) showed that none of the SNPs that mapped to AK5 and HCRTR2 in the current study were in LD at r 2 > .5 with those SNPs associated in previous GWAS (notably, this was limited to SNPs that were in the 1000 Genomes Pilot 1 data and only evaluated LD for subjects of Northern and Western European (CEU) ancestry; thus evaluation of LD proxies for some SNPs was impossible, particularly the previously associated SNPs in FBXL13 as those were not in the 1000 Genomes Pilot 1 data, and LD estimations for our admixed population may be different regardless).
Genetic Risk Score Profiling
Cross-phenotype GRS analysis provided evidence that variants associated with BP and DS nominally predicted some of the risk for being an owl (BP: pT = 0.03, z = −2.505, p = .012; and DS: pT = .49, z = −2.148, p = .032). These results do not survive correction for multiple testing at p < .001. Variants associated with SCZ or BMI did not predict being an owl at p < .05. Plots showing the number of SNPs used in generating each GRS given the p value cutoff are shown in Figure S3.
Discussion
We present the first investigation of genetic risk factors for evening preference in a minority sample of AIs and MAs. Over the last decade several epidemiological studies have reported poorer quality sleep and a higher prevalence of short and/or long sleep in non-White adults, particularly from lower socioeconomic status groups, when compared to White adults (Carnethon et al., Reference Carnethon, De Chavez, Zee, Kim, Liu, Goldberger and Knutson2016; Hale & Do, Reference Hale and Do2007; Krueger & Friedman, Reference Krueger and Friedman2009; Stamatakis et al., Reference Stamatakis, Kaplan and Roberts2007). While not a debilitating disease in itself, evening preference has been associated with several psychiatric disorders. Previous GWAS on chronotype have included predominantly European American samples; two minority samples were used in this study, for whom substance use, anxiety, and depressive disorders have been shown to differentially affect various aspects of sleep (Ehlers et al., Reference Ehlers, Gilder, Criado and Caetano2010; Reference Ehlers, Wills, Lau and Gilder2017). Specifically, in the AI sample the global score on the PSQI was significantly higher in those participants with a lifetime diagnosis of substance use disorders, anxiety disorders, and affective disorders (Ehlers et al., Reference Ehlers, Wills, Lau and Gilder2017), and in the MA sample lifetime diagnoses of alcohol-use disorders, family history of alcohol dependence, acculturation stress, and lifetime diagnoses of major depressive disorder were all correlated with significantly poorer quality sleep as indexed by the global score on the PSQI (Ehlers et al., Reference Ehlers, Gilder, Criado and Caetano2010). The results presented herein suggest that variants previously associated with BP confer risk for being an owl in this minority sample of AIs and MAs.
The primary finding from the GWAS was a suggestively significant signal in KIAA1549L associated with being an owl. One cohort did not drive this association as post hoc analyses indicated the top SNP was associated at p < .05 in the same direction in both cohorts. The KIAA1549L protein, spanning 1,849 amino acids, contains highly conserved regions and had been shown to act as a fusion partner of paired box 5 (PAX5), a transcription factor that is critical for B-cell activation and maintenance (Anderl et al., Reference Anderl, Konig, Attarbaschi and Strehl2015; Medvedovic et al., Reference Medvedovic, Ebert, Tagoh and Busslinger2011). PAX5 was one of the most associated genes with chronotype in a gene-based test from Lane and colleagues’ (Reference Lane, Vlasac, Anderson, Kyle, Dixon, Bechtold and Saxena2016) study of the UK Biobank sample. KIAA1549L has previously been associated with attempted suicide in bipolar patients; in a meta-analysis of a discovery sample (1,201 suicide attempters and 1,497 non-attempters) and replication sample (1,295 attempters and 1,822 non-attempters), a signal in C11orf41 (aka KIAA1549L) was associated at p = 3.77E-06 (Willour et al., Reference Willour, Seifuddin, Mahon, Jancic, Pirooznia, Steele and Potash2012). Although this SNP, rs10437629, was not significantly associated with being an owl in the current study (p = .663; results not shown), the discovery sample used by Willour et al. (Reference Willour, Seifuddin, Mahon, Jancic, Pirooznia, Steele and Potash2012) was of European ancestry. It is likely that neither the signal identified in our study nor the signal identified by Willour and colleagues is the causal signal and, furthermore, that LD patterns between each signal differ between populations. These factors could explain the lack of agreement across studies. Suicidal ideation was positively correlated with evening tendencies in a sample of patients with depression (Bahk et al., Reference Bahk, Han and Lee2014), and given the robust link between evening preference and BD (Ahn et al., Reference Ahn, Chang, Joo, Kim, Lee and Kim2008; Giglio et al., Reference Giglio, Magalhaes, Andersen, Walz, Jakobson and Kapczinski2010; Jeong Jeong et al., Reference Jeong Jeong, Moon, Min Park, Dae Lee, Min Lee, Choi and In Chung2015; Mansour et al., Reference Mansour, Wood, Chowdari, Dayal, Thase, Kupfer and Nimgaonkar2005; Melo et al., Reference Melo, Abreu, Linhares Neto, de Bruin and de Bruin2016; Seleem et al., Reference Seleem, Merranko, Goldstein, Goldstein, Axelson, Brent and Birmaher2015; Wood et al., Reference Wood, Birmaher, Axelson, Ehmann, Kalas, Monk and Nimgainkar2009), this is a prime gene for follow-up studies. Finally, although not suggestively significant, the second two most associated genes in our GWAS, sushi, von Willebrand factor type A, EGF and pentraxin domain containing 1 (SVEP1) and sidekick cell adhesion molecule 2 (SDK2), were nominally associated in previous GWAS with BD and panic disorder, respectively (Ferreira et al., Reference Ferreira, O'Donovan, Meng, Jones, Ruderfer and Jones2008; Otowa et al., Reference Otowa, Yoshida, Sugaya, Yasuda, Nishimura, Inoue and Okazaki2009).
Tangentially, the F-box protein gene FBXO3 is downstream of KIAA1549L (Figure S2). The F-box proteins constitute one of the four subunits of the SKP1-cullin-F-box ubiquitin protein ligase complex which functions in phosphorylation-dependent ubiquitination. This is notable in that previous GWAS have identified SNPs in FBXL13 and FBXL3 (Hu et al., Reference Hu, Shmygelska, Tran, Eriksson, Tung and Hinds2016; Jones et al., Reference Jones, Tyrrell, Wood, Beaumont, Ruth, Tuke and Weedon2016; Lane et al., Reference Lane, Vlasac, Anderson, Kyle, Dixon, Bechtold and Saxena2016) associated with chronotype, thus highlighting the potential role of F-box proteins in diurnal preference.
Although a full investigation of the variants previously associated with chronotype was not possible given the nature of these data, overall there was limited convergence in results between this study and previous GWAS. There are a number of potential explanations for this phenomenon. First, our owl phenotype was loosely defined; many previous studies have used the Horne–Östberg Morningness–Eveningness Questionnaire (Horne & Ostberg, Reference Horne and Ostberg1976), or adaptations of it, to assess diurnal preference. Our case/control phenotype also differed from recent large-scale GWAS that assessed chronotype a number of ways, including using extreme owls as compared to extreme larks and creating a continuous score of diurnal preference (Hu et al., Reference Hu, Shmygelska, Tran, Eriksson, Tung and Hinds2016; Jones et al., Reference Jones, Tyrrell, Wood, Beaumont, Ruth, Tuke and Weedon2016; Lane et al., Reference Lane, Vlasac, Anderson, Kyle, Dixon, Bechtold and Saxena2016). These GWAS were limited to subjects of European ancestry; differences in LD and MAF between ethnicities could further account for discrepancies. Finally, this study was not fully powered to investigate genetic influences, particularly those of small effect, on being an owl, which may have led to false negatives.
Cross-phenotype GRS profiling suggested that being a night owl shares some genetic risk with BP and depression, while no significant GRSs from SCZ or BMI predicted being an owl in this population. Previous work has found no evidence for a genetic correlation between chronotype and BP (Jones et al., Reference Jones, Tyrrell, Wood, Beaumont, Ruth, Tuke and Weedon2016; Lane et al., Reference Lane, Vlasac, Anderson, Kyle, Dixon, Bechtold and Saxena2016). Although it did not survive correction for multiple testing, a GRS from pT = 0.03 indicated only those variants more highly associated with BP predict being an owl in our data, rather than an overall genetic correlation. Conversely, a GRS from the much higher pT = 0.49 using summary statistics on DS nominally predicted being an owl in the MA/AI sample. This is roughly in agreement with previous work: a twin study previously demonstrated a significant genetic correlation of −0.21 between diurnal preference and depression (Toomey et al., Reference Toomey, Panizzon, Kremen, Franz and Lyons2015); and GWAS in the UK Biobank found nominally significant genetic correlations between major depressive disorder and chronotype (Jones et al., Reference Jones, Tyrrell, Wood, Beaumont, Ruth, Tuke and Weedon2016; Lane et al., Reference Lane, Vlasac, Anderson, Kyle, Dixon, Bechtold and Saxena2016), although neither of those p values, nor our own, survived correction for multiple testing.
The UK Biobank GWAS also found significant positive genetic correlations between eveningness and SCZ, and nominal genetic correlations between chronotype and BMI that were lower in magnitude; these findings were not replicated in this study. It is not clear what these negative findings mean, as these results may reflect underlying genetic architecture between populations and environmental factors. With the exception of the summary data on BMI, which was from a multi-ethnic sample, all GWAS summary statistics were derived from European populations. While these negative results may in part be due to the factors listed above (e.g., phenotype definition, limited power, and limited number of imputed variants), the predictive ability of GRSs are sensitive to differences in LD and MAF across base and target samples and previous work on our AI sample indicated that GRSs for smoking behaviors from a European population were only predictive in our sample for those subjects with a high proportion of European ancestry (Otto et al., Reference Otto, Gizer, Bizon, Wilhelmsen and Ehlers2016). The findings presented above highlight the need for both replication and large-scale studies of these minority populations to probe for ethnic-specific genetic risk factors and endophenotypes for complex diseases.
This study provides evidence that genetic risk factors for BP also confer risk to being an owl in a sample of MAs and AIs, suggesting that evening preference may be a useful endophenotype for future studies of BP. This study was underpowered to fully evaluate the phenotype given the small sample size but a signal in KIAA1549L, a gene previously associated with attempted suicide in bipolar patients, was associated with being an owl in both cohorts and, furthermore, a GRS derived from a large-scale meta-analysis of BP nominally predicted being an owl at p = .012. These findings are in line with a body of literature supporting a link between BP and evening preference (Ahn et al., Reference Ahn, Chang, Joo, Kim, Lee and Kim2008; Giglio et al., Reference Giglio, Magalhaes, Andersen, Walz, Jakobson and Kapczinski2010; Jeong Jeong et al., Reference Jeong Jeong, Moon, Min Park, Dae Lee, Min Lee, Choi and In Chung2015; Mansour et al., Reference Mansour, Wood, Chowdari, Dayal, Thase, Kupfer and Nimgaonkar2005; Melo et al., Reference Melo, Abreu, Linhares Neto, de Bruin and de Bruin2016; Seleem et al., Reference Seleem, Merranko, Goldstein, Goldstein, Axelson, Brent and Birmaher2015; Wood et al., Reference Wood, Birmaher, Axelson, Ehmann, Kalas, Monk and Nimgainkar2009). Although these results warrant replication, the growing body of literature exploring the potential overlap between health disparities research and sleep medicine (Jean-Louis & Grandner, Reference Jean-Louis and Grandner2016) and lack of a potential replication sample comprised of MAs and/or AIs simultaneously highlights the importance of conducting studies on these understudied populations.
Acknowledgments
The authors would like to thank: Drs Chris Bizon, Scott Chase, and Jeffrey Tilson for their genotyping efforts; Derek Willis, Dr Qian Peng, and Dr Trina Norden-Krichmar for assistance with data organization and analysis; and Dr David Gilder, Evie Philips, Gina Stouffer, and Corinne Kim for phenotypic data collection. This research was supported by grants from the National Institute on Alcoholism and Alcohol Abuse (NIAAA; grants AA010201 and AA006420 to C.L.E. and AA013525 to E.P.R.) and the National Institute on Drug Abuse (NIDA; grant DA030976 to K.C.W. and C.L.E.) of the National Institutes of Health. Neither NIAAA nor NIDA had any role in the study design, collection, analysis, or interpretation of the data, nor in the writing and submission of this work.
Conflict of Interest
None.
Supplementary material
To view supplementary material for this article, please visit https://doi.org/10.1017/thg.2017.62





