Genetic differences account for a significant proportion of neuroanatomic variability in humans (Hulshoff Pol et al., Reference Hulshoff Pol, Schnack, Posthuma, Mandl, Baare, van Oel, van Haren, Collins, Evans, Amunts, Bürgel, Zilles, de Geus, Boomsma and Kahn2006; Pennington et al., Reference Pennington, Filipek, Lefly, Chhabildas, Kennedy, Simon, Filley, Galaburda and DeFries2000; Pfefferbaum et al., Reference Pfefferbaum, Sullivan, Swan and Carmella2000). While several studies have considered heritable influences on total brain volume (Cheverud et al., Reference Cheverud, Falk, Vannier, Konigsberg, Helmkamp and Hildebolt1990; Posthuma et al., Reference Posthuma, de Geus, Baare, Hulshoff Pol, Kahn and Boomsma2002; Rogers et al., Reference Rogers, Kochunov, Lancaster, Shelledy, Glahn, Blangero and Fox2007; Rogers et al., Reference Rogers, Kochunov, Zilles, Shelledy, Lancaster, Thompson, Duggirala, Blangero, Fox and Glahn2010; Thompson et al., Reference Thompson, Cannon, Narr, van Erp, Poutanen, Huttunen, Lönnqvist, Standertskjöld-Nordenstam, Kaprio, Khaledy, Dail, Zoumalan and Toga2001; Toga & Thompson, Reference Toga and Thompson2005), little is known about genetic influences on regional structures such as the corpus callosum (CC). The CC is the largest commissural white-matter (WM) tract in the brain, and is essential for inter-hemispheric integration of sensory, motor, and higher-order cognitive information. Numerous genetic disorders affect the morphology of the CC, producing specific regional abnormalities (Di Rocco et al., Reference Di Rocco, Biancheri, Rossi, Filocamo and Torotori-Donati2004; Kochunov et al., Reference Kochunov, Lancaster, Hardies, Thompson, Woods, Cody, Hale, Laird and Fox2005). Disruptions in the structural integrity of the CC during aging, or as a result of specific disorders, are associated with impairments in problem-solving and working memory (Zahr et al., Reference Zahr, Rohlfing, Pfefferbaum and Sullivan2009), bimanual movement, or inter-hemispheric transfer (Bonzano et al., Reference Bonzano, Tacchino, Roccatagliata, Abbruzzese, Mancardi and Bove2008). Neuropsychiatric conditions, including schizophrenia (Wang et al., Reference Wang, Deng, Huang, Li, Ma and Wang2011) and major depression (Korgaonkar et al., Reference Korgaonkar, Grieve, Koslow, Gabrieli, Gordon and Williams2011), are associated with changes in the CC. Given the importance of the CC to various cognitive functions, understanding the genetic mechanisms that influence variation in the size and shape of this structure will likely have important clinical implications.
Using MRI, human twin studies have suggested a high heritability of the midsagittal CC area (Scamvougeras et al., Reference Scamvougeras, Kigar, Jones, Weinberger and Witelson2003). Analyses in a small number of mono- (N = 10) and dizygotic (N = 7) twin pairs estimated heritability at 94.4% for the size of the CC. Pfefferbaum et al. (Reference Pfefferbaum, Sullivan, Swan and Carmella2000) reported similarly high heritability (85%) for the CC size in another small (N = 85) twin sample. More recent investigations into the regional heritability of the CC partitions using diffusion tensor imaging (DTI) have reported that the degree of contributions by genetic factors was variable among midsagittal CC sections, and the sources of this variability remained unknown (Brouwer et al., Reference Brouwer, Mandl, Peper, van Baal, Kahn, Boomsma and Hulshoff Pol2010; Chiang et al., Reference Chiang, McMahon, de Zubicaray, Martin, Hickie, Toga, Wright and Thompson2011; Kochunov et al., Reference Kochunov, Glahn, Lancaster, Wincker, Smith, Thompson, Almasy, Duggirala, Fox and Blangero2010c). Some developmental biologists have suggested that the rate of development may modulate the degree of genetic contribution, and earlier developing structures will be more tightly controlled by genetic factors during development, thus leading to higher heritability. However, this assertion is not consistent with results that demonstrate that the earliest developing structures are clearly evolvable through the course of the evolutionary history, and may be susceptible to environmental perturbations (Raff, Reference Raff1996). This was further demonstrated by recent studies that showed that the heritability of WM increases with age (Peper et al., Reference Peper, Brouwer, Boomsma, Kahn and Hulshoff Pol2007), and the brain regions associated with more complex reasoning become increasingly more heritable with development (Lenroot & Giedd, Reference Lenroot and Giedd2008).
Comparative studies of animal models can provide unique insights into the biological processes that underlie human neurobiology and neurodevelopment. Previous studies of baboons have documented significant genetic effects on brain structure (Kochunov et al., Reference Kochunov, Glahn, Fox, Lancaster, Saleem, Shelledy, Zilles, Thompson, Coulon, Mangin, Blangero and Rogers2010b; Rogers et al., Reference Rogers, Kochunov, Lancaster, Shelledy, Glahn, Blangero and Fox2007), and shown that there are unanticipated parallels in the architecture of genetic effects on cortical folding and brain volume in humans and baboons (Rogers et al., Reference Rogers, Kochunov, Zilles, Shelledy, Lancaster, Thompson, Duggirala, Blangero, Fox and Glahn2010). In this latter paper, we showed that an inverse relationship between genetic effects on brain size and cortical folding is conserved in humans and baboons. Thus, the baboon results provide both a confirmation of an unexpected finding in human neurogenetics and demonstrate that there are long-term evolutionary genetic relationships that are shared across primate clades. Significant results concerning brain structure and the genetics of brain evolution have also come from studies of macaques, vervet monkeys, and chimpanzees (e.g., Fears et al., Reference Fears, Melega, Service, Lee, Chen, Tu, Jorgensen, Fairbanks, Cantor, Freimer and Woods2009; Lyn et al., Reference Lyn, Pierre, Bennett, Fears, Woods and Hopkins2011; Semendeferi et al., Reference Semendeferi, Lu, Schenker and Damasio2002; Sherwood et al., Reference Sherwood, Raghanti, Stimpson, Spocter, Uddin, Boddy, Wildman, Bonar, Lewandowski, Phillips, Erwin and Hof2010). In addition to comparative analyses of nonhuman primate and human brain structure, researchers have also successfully used nonhuman primates to study genetic influences on brain function and metabolism (Oler et al., Reference Oler, Fox, Shelton, Rogers, Dyer, Davidson, Shelledy, Oakes, Blangero and Kalin2010). Between-species differences in gene expression within the brain have also informed our understanding of human brain function (Konopka et al., Reference Konopka, Bomar, Winden, Coppola, Jonsson, Gao, Peng, Preuss, Wohlschlegel and Geschwind2009).
We aimed to evaluate genetic influences on inter-subject variability in midsagittal CC size, and the degree to which the genetic heritability of regional CC variability was modulated by rate of development during fetal and early postnatal growth. The evaluation was performed in a nonhuman primate: baboons, Papio hamadryas. Papio baboons were chosen because they share several neurological characteristics with humans, including high heritability of brain volume, cortical surface area, and cortical gyrification (Kochunov et al., Reference Kochunov, Glahn, Fox, Lancaster, Saleem, Shelledy, Zilles, Thompson, Coulon, Mangin, Blangero and Rogers2010b). Additionally, age-related changes in the development of the CC are consistent with the developmental course observed in humans (Phillips & Kochunov, 2011). Therefore, the baboon holds great potential as a model for human brain development.
Methods
Subjects
One hundred-eighty adult baboons (Papio hamadryas) (68 males, 112 females) were selected from the large multi-generation pedigreed colony of more than 2000 baboons maintained by the Southwest National Primate Research Center (SNPRC) at the Texas Biomedical Research Institute in San Antonio, Texas. The average age of the study animals was 16 years (SD = 4.2, age range: 7–28 years). This age range was chosen to minimize the effects of development or senescence based on studies of cerebral ontogeny (Leigh, Reference Leigh2004; Leigh et al., Reference Leigh, Shah and Buchanan2003). The genealogical relationships among study animals included 414 parent–offspring pairs, 51 full sib pairs, 645 half-sib pairs, and a large number of more distant kinship relationships. Captive male baboons are sexually mature at 5 years and fully adult at 6 years. Female baboons start to cycle at between 3–4 years and are fully grown around 5 years.
We measured the CC in utero and during the early postnatal period to estimate developmental rate. In-utero imaging of 13 normally developing fetuses was performed covering the period of gestational week 17 through birth (gestational week 28); postnatal imaging was performed on 16 baboons between postnatal weeks 1 and 32. The details for the in-utero and early postnatal imaging and animal handling protocols are described elsewhere (Kochunov et al., Reference Kochunov, Castro, Davis, Dudley, Brewer, Zhang, Kroenke, Purdy, Fox, Simerly and Schatten2010a; Kochunov & Duff Davis, Reference Kochunov and Duff Davis2009; Phillips & Kochunov, 2011).
Animal Handling and MR Imaging
Animals were transported from the SNPRC to the Research Imaging Institute, University of Texas Health Sciences Center at San Antonio for imaging. Handling and anesthesia procedures followed procedures described previously (Kochunov & Duff Davis, 2009; Rogers et al., Reference Rogers, Kochunov, Lancaster, Shelledy, Glahn, Blangero and Fox2007); they are briefly summarized here. Fifteen minutes prior to scanning, animals were immobilized with ketamine (10 mg/kg) and intubated with an MR-compatible endotracheal tube. Anesthesia was maintained with 5% isoflurane with an MR-compatible gas anesthesia machine. Animals remained anesthetized throughout the imaging procedure; respiration rate, heart rate, and oxygen consumption were monitored continuously. This protocol and all animal procedures were reviewed and approved by the Institutional Animal Care and Use Committee of the Texas Biomedical Research Center.
The imaging protocols used to acquire images from all subjects are detailed elsewhere (Kochunov & Duff Davis, 2009; Rogers et al., Reference Rogers, Kochunov, Lancaster, Shelledy, Glahn, Blangero and Fox2007). In short, high-resolution (isotropic 500 μm), T1-weighted images were acquired using a 3D IR-TurboFlash sequence optimized for anatomical imaging of baboon brain. An adiabatic inversion recovery (IR) contrast pulse with linear phase encoding schema was employed, primarily because it led to a uniform tissue contrast across the imaging volume (being less affected by B1-inhomogeneity/radio-frequency [RF] penetration artifacts). The sequence control parameters for the adult and postnatal subjects (FOV = 128 mm, TI = 795 ms, TE = 3.04, TR1 = 5 ms, TR2 = 2000 ms, and flip angle = 10°) were modeled to produce gray matter–white matter (GM–WM) contrast of 25% based on the analytical solutions to Bloch equations (Deichmann et al., Reference Deichmann, Good, Josephs, Ashburner and Turner2000), and average measured values of T1, T2, and PD. The model-determined imaging sequence parameters were verified in a group of five animals, where group-average GM–WM contrast was calculated to be 25.2 ± 2% (range 22–26%). Image acquisition was performed using a retrospective motion-corrected protocol (Kochunov et al., Reference Kochunov, Lancaster, Glahn, Purdy, Laird, Gao and Fox2006). Under this protocol, six full-resolution segments, each 9 minutes long, were acquired for a total sequence running time of ~54 minutes. The sequence control parameters, TR/TE/flip angle/FOV/Spatial Resolution/Scan Time = 5 ms/2.5 ms/75 degrees/180 mm/500 microns isotropic/30 min, allow for rapid collection of 3D data phase partition of 360 lines within a single respiration cycle, as detailed in Phillips & Kochunov, 2011. This protocol allowed for a high-SNR, 3D, and isotropic coverage of the fetal brain, with good regional GM–WM tissue contrast.
Image Processing and Measurement of CC
The image processing pipeline consisted of the following steps: removal of non-brain tissue, correction for spatial variations in intensity due to scanner radio-frequency inhomogeneity, and global spatial normalization to a population-based template to reduce global variability in brain size and orientation (Figure 1). The details of this processing are described elsewhere (Rogers et al., Reference Rogers, Kochunov, Lancaster, Shelledy, Glahn, Blangero and Fox2007). In short, the removal of non-brain tissue used both automatic (Smith, Reference Smith2002) and manual detailing methods. The correction for RF-inhomogeneity was performed using the functional magnetic resonance imaging of the brain (FMRIB) automated segmentation tool (Smith et al., Reference Smith, Jenkinson, Woolrich, Beckmann, Behrens, Johansen-Berg, Bannister, De Luca, Drobnjak, Flitney, Niazy, Saunders, Vickers, Zhang, De Stefano, Brady and Matthews2004). A nine-parameter global spatial normalization procedure was used to reduce inter-subject variability in global brain size, shape, and orientation, and was performed using the FMRIB linear image registration tool (Smith et al., Reference Smith, Jenkinson, Woolrich, Beckmann, Behrens, Johansen-Berg, Bannister, De Luca, Drobnjak, Flitney, Niazy, Saunders, Vickers, Zhang, De Stefano, Brady and Matthews2004). A population-based, pseudo-Talairach, median-geometry atlas served as the target brain for global spatial normalization. This atlas was created using methods previously described for humans (Kochunov et al., Reference Kochunov, Lancaster, Thompson, Toga, Brewer, Hardies and Fox2002) and primates (Kochunov & Duff Davis, 2009).
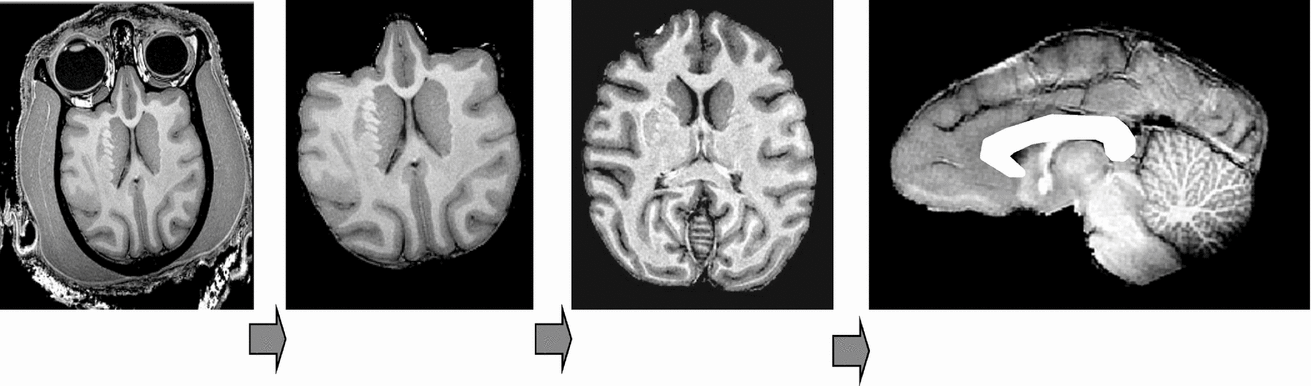
FIGURE 1 Structural image processing pipeline, which allows for a simple automation of sequential processing steps. Our pipeline consists of the following steps: removal of non-brain tissue, correction for RF-inhomogeneity artifacts, global spatial normalization (A), hemispheric segmentation (B), tissue classification (C), extraction of the inner/outer cortical surfaces (D, E), extraction of cortical sulci (F), automated labeling of cortical sulci (G), and gyral segmentation (H).
Measurements of CC area were then performed from the midsagittal section, where the CC can be readily identified, using methodology originally described by Biegon and colleagues (Biegon et al., Reference Biegon, Eberling, Richardson, Roos, Wong, Reed and Jagust1994), and later adapted to nonhuman primates (Sanchez et al., Reference Sanchez, Hearn, Do, Rilling and Herndon1998). In the original procedure, the anterior 20% of the CC was defined as the genu, the posterior 20% defined as the splenium, and the middle 60% defined as the body. In adapting this to nonhuman primates, Sanchez et al. (Reference Sanchez, Hearn, Do, Rilling and Herndon1998), Phillips et al. (Reference Phillips, Sherwood and Lilak2007), and Pierre et al. (Reference Pierre, Hopkins, Taglialatela, Lees and Bennett2008), further delineated the body into three equal regions: anterior midbody, medial midbody, and caudal midbody. These subdivisions of the CC are believed to correspond to functional connectivity with cortical areas (Aboitiz et al., Reference Aboitiz, Scheibel, Fisher and Zaidel1992; Alexander et al., Reference Alexander, Lee, Lazar, Boudos, Dubray, Oakes, Miller, Lu, Jeong, McMahon, Bigler and Lainhart2007; Hofer & Frahm, Reference Hofer and Frahm2006). The anterior region of the genu and anterior midbody connects higher-association areas of the frontal lobe; the medial and caudal midbody connect primarily sensorimotor regions; the posterior region of the splenium integrates visuospatial regions of the cortex. Analyze 10.0 (Mayo Foundation for Medical Education and Research) was used to divide and measure the midsagittal area of the CC in mm2. To subdivide the CC, the entire length of the CC was first manually traced, then divided into five equally spaced sections (see Figure 2). Two individuals (KAP and EAB) performed measurements of the CC; there was a high degree of concordance in measures, r = .88. Details on the measurements of total CC area and CC subdivision area were provided in Phillips and Kochunov (Reference Phillips and Kochunov2011). Regional development rates were estimated by fitting a linear regression to the dataset consisting of both in-utero and postnatal data points. In this way we estimated mm2/week of development in callosal subdivisions.

FIGURE 2 Anatomical subdivision of the baboon corpus callosum from MRI sagittal view. The total midsagittal area was divided into five equally spaced subdivisions. 1 = genu; 2 = anterior midbody; 3 = medial midbody; 4 = caudal midbody; 5 = splenium.
Quantitative Genetic Analysis
Variance components methods, as implemented in the SOLAR software package (http://solar.sfbrgenetics.org) (Almasy & Blangero, Reference Almasy and Blangero1998), were used to estimate the heritability of measured traits. The algorithms in SOLAR employ maximum likelihood variance decomposition methods and are an extension of the strategy developed by Amos (Reference Amos1994). The covariance matrix Ω for a pedigree of individuals is given by Equation 1:
where s2g is the genetic variance due to the additive genetic factors, F is the kinship matrix representing the pair-wise kinship coefficients among all animals, s2e is the variance due to individual‑specific environmental effects, and I is an identity matrix. The kinship matrix F was calculated based on the known breeding records and was verified by genetic microsatellite-marker-based testing that confirmed parent–offspring relationships among baboons. This produces a multigenerational pedigree that summarizes the genetic relationships among all individuals. For additional explanation of the variance components approach in this context, see Almasy & Blangero (Reference Almasy and Blangero1998) and Blangero et al. (Reference Blangero, Williams and Almasy2001).
Heritability (h 2), the portion of phenotypic variance (sp2) that is accounted for by additive genetic variance (Eq. 1), is assessed by contrasting the observed phenotypic covariance matrix with the covariance matrix predicted by kinship. Significance of heritability is tested by comparing the likelihood of the model in which s2g is constrained to zero with that of a model in which s2g is estimated. Twice the difference between the two log likelihoods of these models yields a test statistic which is asymptotically distributed as a ½:½ mixture of a χ12 variable and a point mass at zero. During testing for the significance of heritability, the phenotype values for each individual are adjusted for a series of covariates. In our analysis we used a polygenic model that estimated the influence of specific variables (additive genetic variation and covariates including sex, age, age2, age x sex interaction, age2 x sex interaction, and random unidentified environmental effects), calculating heritability and its significance (p value) for each trait's variance within this population. The level of significance for the heritability analysis for callosal subdivisions was set at p ≤ .01 (Bonferroni correction) to reduce the probability of Type 1 errors associated with multiple (N = 5) measurements.
Genetic Correlation Analyses
Bivariate genetic correlation analyses were performed to study the proportion of shared genetic variance between the subdivisions of the CC using methods implemented in the SOLAR software package. Bivariate genetic analysis calculates the magnitude and significance of genetic correlation coefficient (ρ G), which is the proportion of variability due to shared genetic effects. The overall phenotypic correlation (ρ P) between two traits A and B (Equation 2) can be expressed using the correlation due to shared additive genetic effects (ρ G), and the residual correlation (ρ E) due to shared environmental effects.
where h2A and h2B denote the additive genetic heritabilities for each of the traits, that is, the proportion of the total phenotypic variance that is explained by additive genetic factors. If the genetic correlation coefficient (ρ G) is significantly different from zero, then the traits are considered to be partially influenced by shared genetic factors (Almasy et al., Reference Almasy, Dyer and Blangero1997).
Results
The mean area for callosal subdivisions in adult baboons is shown in Table 1. Quantitative genetic analyses revealed that the total area of the CC and all subdivisions is heritable, with h 2 at .46 (SD = .16) for the total CC, ranging from .29 (SD = .14) for the splenium to .62 (SD = .17) for the medial midbody (Table 1). Bivariate genetic correlation analysis demonstrated that the individual subdivisions shared between 41% and 98% of genetic variability (Table 2). There were no significant covariates for any of the phenotypes, which was likely a result of using global spatial normalization to correct for differences in head size.
TABLE 1 Mean Area, Rates of Development, Heritability, and Proportion of the Total Variance Explained by Covariates for the Corpus Callosum and Regional Subdivisions in Adult Baboons

CC = corpus callosum, SD = standard deviation.
a Data from Phillips and Kochunov (Reference Phillips and Kochunov2011).
TABLE 2 Genetic Correlations Between the Subdivisions of the Corpus Callosum
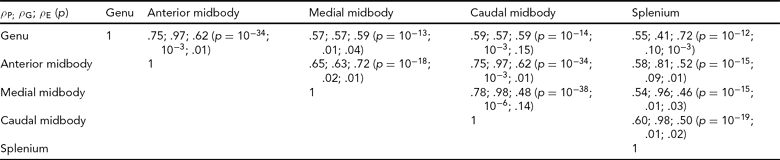
The overall phenotypic correlation (ρP) between two traits is expressed using the correlation due to shared additive genetic effects (ρG) and the residual correlation (ρE) due to shared environmental effects.
To test if the rate of cerebral development was predictive of the level of heritability in adulthood, we plotted the degree of genetic contribution to regional variability (i.e., heritability in size) in the CC subdivisions versus the regional development rates during the fetal and early postnatal period. These rates were determined from brain images of 29 normally developing fetuses, covering the period of gestational week 17 through birth (gestational week 28). Imaging was also performed on 16 baboons up to postnatal week 32. The regional estimates of heritability were negatively correlated with the rates of change in midsagittal CC area during this developmental period (r = .74, p = .08) (Table 1).
Discussion
Our study demonstrated significant heritability for the size of the CC and its subdivisions in adult baboons. To our knowledge, this is the first investigation of heritability of the CC subdivisions performed in a pedigree of nonhuman primates. Heritability of individual callosal subdivisions varied from .29 (SD = .14) for the splenium to .62 (SD = .17) for the medial midbody, suggesting callosal subdivisions may differ in the degree of genetic contribution to the individual variation, but the large standard errors we obtained suggest that in this dataset the heritabilities are not statistically different. Overall, the estimates of heritability in baboons were about half of those reported in humans (Pfefferbaum et al., Reference Pfefferbaum, Sullivan, Swan and Carmella2000; Scamvougeras et al., Reference Scamvougeras, Kigar, Jones, Weinberger and Witelson2003). This discrepancy can potentially be explained by the methodological differences, since the human studies did not correct for inter-subject differences in brain volume. The total brain volume is highly heritable in both humans and baboons (Rogers et al., Reference Rogers, Kochunov, Lancaster, Shelledy, Glahn, Blangero and Fox2007; Rogers et al., Reference Rogers, Kochunov, Zilles, Shelledy, Lancaster, Thompson, Duggirala, Blangero, Fox and Glahn2010), and this study aimed to measure genetic contribution of the morphology of CC independent of brain size. The global differences in brain size and shape can be corrected by spatial normalization, which transforms brains into a standard reference frame using a nine-parameter (three translations, rotations, and scaling) spatial transformation where they are adjusted to the same external dimensions (Rogers et al., Reference Rogers, Kochunov, Lancaster, Shelledy, Glahn, Blangero and Fox2007). This normalization step was also shown to remove the effects of body weight and sex (Kochunov et al., 2009; Rogers et al., Reference Rogers, Kochunov, Lancaster, Shelledy, Glahn, Blangero and Fox2007). After spatial normalization, variability in brain structure chiefly reflects individual variability in the structure's shape, such as curvature, length, and width. Additionally, the investigations of CC heritability in humans were performed on a small number of twin pairs and used a simplified estimate of heritability that did not model the shared environmental effects, which are known for overestimation of the heritability values (Keller et al., Reference Keller, Medland and Duncan2010).
As brain regions associated with more complex reasoning were reported to become increasingly heritable with maturation (Lenroot & Giedd, Reference Lenroot and Giedd2008), we expected higher heritability in subdivisions of the CC connecting higher-association areas. The subdivisions of the CC are associated with functional connectivity to cortical regions (Alexander et al., Reference Alexander, Lee, Lazar, Boudos, Dubray, Oakes, Miller, Lu, Jeong, McMahon, Bigler and Lainhart2007; Hofer & Frahm, Reference Hofer and Frahm2006). The anterior regions of the genu and anterior midbody connect primarily higher-order cognitive regions; the medial and caudal midbody connect primarily sensorimotor regions; the posterior region of the splenium integrates visuospatial regions of the cortex. Thus, as the genu and splenium are involved in higher-association cognitive tasks, we expected these subdivisions to have the greatest heritability in adult baboons. When considering heritability of the CC in adults, the genu and splenium did not display the highest heritability rates. The value for the genu, which connects prefrontal regions, is higher than the average across regions, and higher than the value for the total CC, but the value of heritability for the splenium is lower than all other values. As the splenium is involved in visuospatial integration, perhaps experiences in coordination of visual and motor activities are important factors in influencing the variability in size of the splenium. When looking at heritability rates across fetal and early postnatal development, a similar pattern is seen. The splenium showed the largest degree of variation due to environmental effects. The results of the genetic correlation analyses revealed that the degree of shared genetic contribution among CC subdivisions is complex, and suggested that subdivisions may differ in the degree of genetic contribution. Heritability among subdivisions did not vary along the anterior–posterior direction, but spatially adjacent subdivisions shared more genetic variability than more distal regions.
The second aim in this study was to investigate whether genetic contributions to inter-subject variability were modulated by the rate of development. Previously we calculated the regional rates of increase in midsagittal CC areas from fetal and early postnatal baboons (Phillips & Kochunov, 2011). Our findings of negative correlation between regional heritability values and the rate of development may suggest that the genetic contributions to regional CC size are negatively correlated with rate of development. Cheverud and colleagues (1990) drew a connection between developmental factors, such as prenatal neurohormonal environment, and the genetic versus environmental contributions to variability in the length of cortical sulci. They found lower heritability estimates for the length of the primary sulci that appear later in cerebral development (Cheverud et al., Reference Cheverud, Falk, Vannier, Konigsberg, Helmkamp and Hildebolt1990). This implied that lower heritability for later-appearing sulci may be due to higher contributions of environmental factors to overall phenotypic variance. They suggested that higher environmental contribution to sulcal morphology could be due to changes in prenatal hormone-mediated neurohumoral environment and tissue receptivity, which become progressively more variable during development (Cheverud et al., Reference Cheverud, Falk, Vannier, Konigsberg, Helmkamp and Hildebolt1990). A trend toward higher heritability values for primary cortical structures appearing earlier in development was also reported in humans (Brun et al., Reference Brun, Lepore, Pennec, Chou, Lee, Barysheva, de Zubicaray, Meredith, McMahon, Wright, Toga and Thompson2008; Chiang et al., Reference Chiang, Barysheva, Lee, Madsen, Klunder, Toga, McMahon, de Zubicaray, Meredith, Wright, Srivastava, Balov and Thompson2008; Le Goualher et al., Reference Le Goualher, Argenti, Duyme, Baare, Hulshoff Pol, Boomsma, Zouaoui, Barillot and Evans2000; Lohmann et al., Reference Lohmann, von Cramon and Colchester2008; Lohmann et al., Reference Lohmann, von Cramon and Steinmetz1999). Our result is supportive of this hypothesis; however, the negative relationship between heritability and the rate of development is driven primarily by the splenium, which had the highest rate of development and the lowest heritability estimate among the CC regions. The value of the correlation coefficient is greatly diminished if splenium is removed from the analysis (r = −.11 vs. −.74, respectively).
Understanding how genes and environmental variation determine brain structure and function is fundamentally important to understanding normal and disease-related patterns of neural structure and function. Significant genetic effects have been reported for other brain structures in baboons, including total brain volume and shape, and regions of motor cortex and the superior temporal gyrus (Rogers et al., Reference Rogers, Kochunov, Lancaster, Shelledy, Glahn, Blangero and Fox2007). Thus, a consistent pattern of high heritability for brain morphometric measures is seen in baboons. The results of the present study further indicate that Papio baboons are a valuable model for translational neurologic genetic research.
Acknowledgments
This work was supported, in part, by the National Institute of Neurological Disorders and Stroke (R15 #NS070717) to KAP, and the National Institute of Biomedical Imaging and Bioengineering (K01 #EB006395) to PK. This investigation used resources that were supported by the Southwest National Primate Research Center grant P51 RR013986 from the National Center for Research Resources, National Institutes of Health and that are currently supported by the Office of Research Infrastructure Programs through P51 OD013986.






