INTRODUCTION
Campylobacter spp. continue to be the leading bacterial cause of infectious intestinal disease in the UK and worldwide. In the UK in 2010, over 70 000 confirmed laboratory cases were reported to the Health Protection Agency (HPA), Centre for Infections [1]. Most infections in humans in the developed world are caused by C. jejuni and C. coli and the majority of these cases are attributed to the consumption of poultry [Reference Studahl and Andersson2–Reference Wingstrand4]. C. jejuni, which causes the highest number of infections in humans, is frequently isolated from poultry and there is no doubt that this meat is a major source of Campylobacter [Reference Jørgensen5]. In a recent EU survey, about 75% of flocks at the time of slaughter and 86% of post-chill carcasses sampled in the UK were contaminated with Campylobacter spp. [6]; they are frequently isolated throughout poultry production, including during rearing and at slaughter [Reference Nebola and Steinhauserova7] and their occurrence is well documented [Reference Wagenaar, Mevius and Havelaar8–Reference Stern10]. However, little is known about the epidemiology of Campylobacter in poultry flocks [Reference Humphrey9], making control measures more difficult to implement and monitor. C. coli can be isolated from pigs, cattle, sheep and wild birds [Reference Humphrey9] but also from poultry [Reference Bull11–Reference Williams13].
Detection of Campylobacter in food, faecal and environmental samples, is performed by either direct plating onto selective agars or by enrichment culture. Isolation from these sample types is considered to be difficult and time-consuming, as campylobacter numbers can be low and other factors such as pH, oxidative stress, competing flora and high incubation temperature inhibit recovery [Reference Verhoeff-Bakkenes14–Reference Yamasaki16]. Selective media were originally developed to isolate Campylobacter from faeces [Reference Corry17]; these media used a mixture of antibiotics in a rich basal medium, which allows these organisms to grow while inhibiting the growth of contaminants. These media can also contain haemin and charcoal as oxygen-quenching agents which can considerably improve recovery rates [Reference Hutchinson and Bolton18, Reference Hunt, Abeyta and Tran19].
There is no universally accepted standard method for isolating Campylobacter spp. [Reference Corry17]. Bolton broth or the modified (mBolton) version of it, which uses amphotericin B instead of cycloheximide, is recommended by the United States Food and Drug Administration (US FDA) [Reference Hunt, Abeyta and Tran19] and The International Standards Organization (ISO 10272-1) [20] and, due to these endorsements, is the most favoured enrichment method.
Alternative enrichment broths are Park & Sanders, and Preston, both of which were previously recommended by the ISO. However, the use of these two broths is decreasing; Preston is used in Australia and recommended as their standard method [21]. There is also Exeter broth, which is commonly used in the UK. A number of studies [Reference Jørgensen5, 6, Reference Josefsen22–Reference Paulsen25] have compared the effectiveness of enrichment broths. Bolton and Preston broths are commonly used media for the isolation of Campylobacter spp. from complex sample types including food, water and those from the environment. Exeter enrichment [Reference Humphrey, Mason and Martin23] has been used successfully to isolate Campylobacter spp. from retail chicken and water samples [6, Reference Josefsen22–Reference Humphrey, Martin and Mason24]. The composition of Exeter broth was altered to use Bolton broth base (Oxoid, UK) creating modified Exeter (mExeter) enrichment broth [Reference Mattick26], which improved the isolation of Campylobacter.
The incubation temperature of enrichment broths can also affect recovery of Campylobacter spp. [Reference Verhoeff-Bakkenes14–Reference Yamasaki16]. Traditionally incubation of these media was performed at 42°C to isolate C. jejuni and C. coli [Reference Scates, Moran and Madden27] but over time the incubation temperature has altered and each enrichment method has a recommended temperature. For example, Exeter broth is recommended to be incubated at 37°C [Reference Mattick26] and Bolton for the first 4 h at 37°C and then at 42°C for 42–44 h; this varies between laboratories and countries and may influence recovery [Reference Scates, Moran and Madden27].
The sampling method and broth used are thus important considerations when selecting an enrichment method [Reference Baylis28, Reference Jørgensen5]. Little is known about whether enrichment methods recover C. jejuni and C. coli and subtypes within each of these species equally well and methods should be free of bias for the subpopulations of different species. Thus, for example, for the identification of sources of infection in poultry flocks it is vital that the methods applied to very different sample types such as faecal and environmental samples allow an equal chance for all Campylobacter types present to be detected.
This study aimed first, to compare the isolation of Campylobacter spp. from poultry faecal and environmental samples using enrichment procedures based on Preston, mBolton and mExeter selective broths, and second, to compare the subtypes of Campylobacter recovered by the three enrichment procedures.
METHODS
Sample preparation
Samples from the chicken-rearing environment were collected from two free-range organic farms in South West England (farms A and B). The flocks sampled were aged between 70 and 80 days. Surfaces (n=93) from the interior and exterior of five broiler houses, including walls, doors, windows, supporting beams, drinkers, air vents, ceilings and perches, were sampled using sterile cloth swabs (Robinsons Health Care, UK). Swabs were placed in a sterile plastic bag and moistened with 10 ml maximum recovery diluent (MRD; Oxoid) immediately prior to use. The swab was held within the bag to reduce contamination and was rubbed over a sample surface of ~0·1 m2 and sealed in the bag. Boot overshoes (n=9) (Mike Bowden Livestock Services, UK), worn throughout sampling in and around individual houses on the farms, were placed into sterile plastic bags for subsequent analysis. Floor material (litter, n=5) from inside the house and the outdoor run, feed (n=5), soil (n=5) and water (n=12) from inside and outside broiler houses were collected in sterile containers. Fresh chicken faecal samples (n=15) were collected in sterile containers. Samples were transported to the laboratory at ambient temperature and analysed within 2 h of collection.
Individual swabs were placed in sterile Stomacher bags using alcohol-sterilized forceps, 10 ml of Nutrient broth no. 2 (NB2; Oxoid) was added and the samples were homogenized using a Lab-Blender 400 for 30 s. Swabs were cut into six pieces using alcohol-sterilized scissors, another 10 ml NB2 was added and homogenized for a further 1 min. Two pieces of the swab to make one sample for enrichment were chosen at random for each broth and placed in sterile 60-ml containers. This method was also used to examine the boot overshoes.
Individual litter, and soil or feed samples were mixed thoroughly using a cotton-tipped swab and 25-g aliquots placed in sterile 250-ml containers for enrichment. Water samples were inverted several times to mix and 25 ml were placed in sterile 250-ml containers. Individual faecal samples were mixed using a cotton-tipped swab and ~2 g placed in sterile 25-ml containers.
Isolation and identification of Campylobacter spp
Three enrichment methods for isolation of Campylobacter spp. were used (Table 1). The broths were added to the sample leaving <1 cm headspace in tightly capped containers and were incubated according to the manufacturer's recommendations for 48 h. After incubation, 10 μl from each broth was streaked onto modified charcoal cefoperazone deoxycholate agar (mCCDA; Oxoid) and these plates were incubated at 37°C for 48 h under microaerobic conditions consisting of 10% CO2, 10% H2 and 80% N2 in Anoxomat jars (Launch Diagnostics Ltd, UK).
Table 1. Enrichment methods for the isolation of Campylobacter spp.
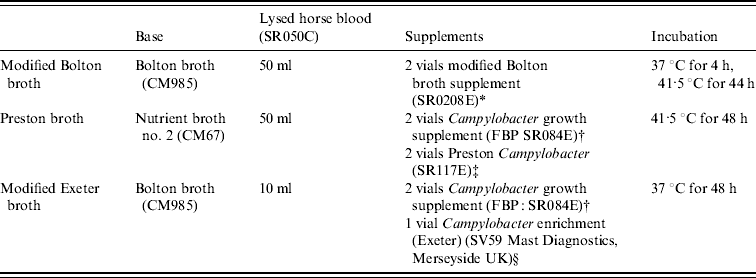
All media and supplements were supplied by Oxoid Ltd, Basingstoke, UK, unless stated otherwise and volumes described are for 1 litre.
* 20 mg cefoperazone, cancomycin and trimethoprim, 10 mg amphotericin B.
† 0·5 g sodium pyruvate, sodium metabisulphite and ferrous sulphate.
‡ 5 mg trimethoprim, 5 mg rifampicin, 2500 IU polymyxin B, 5 mg amphotericin.
§ 10 mg trimethoprim, 5 mg rifampicin, 2500 IU polymyxin B, 15 mg cefoperazone, 2 mg amphotericin.
Presumptive colonies of Campylobacter spp. were confirmed by growth on duplicate plates of Columbia blood agar with 5% (v/v) defibrinated horse blood (COLBA; Oxoid). The two plates were incubated under different conditions at 37°C for 48 h; one aerobically and one microaerobically. Any isolates that were aero-tolerant were presumed not to be Campylobacter spp. and did not undergo further confirmatory tests. Presumptive positives were also confirmed by microscopy and by positive oxidase reaction.
Speciation of Campylobacter isolates
DNA was prepared from the isolates using crude cell lysates in the first instance. For isolates that were not detected by PCR, DNA was extracted using the DNeasy Blood and Tissue kit (Qiagen, UK) on a new subculture of the isolate according to the manufacturer's instructions. Cell lysates were prepared by suspending a 1-μl loop of bacterial growth from a Columbia blood agar plate in 500-μl sterile nuclease-free water (Sigma, UK) and heating the suspension at 100°C for 10 min
Isolates were speciated using a multiplex PCR containing two primer sets for each species (C. jejuni and C. coli) referred to as multiplex PCR throughout (Table 2). Primers were used at 10 μm concentrations and each PCR reaction contained 0·5 μl of each primer, 12·5 μl Hotstart Taq Mastermix (Qiagen), 0·75 μl of 50 mm MgCl2, 2·75 μl nuclease-free water and 5 μl template DNA. Cycling parameters were as follows; 95°C for 15 min, 30 cycles of 94°C for 1 min, 60°C for 1 min, 72°C for 1 min with a final extension time of 72°C for 10 min.
Table 2. Primers used in the hipO/lpxA/glyA multiplex PCR for the identification of C. jejuni and C. coli

DNA electrophoresis was performed with a Mini-Sub Cell GT (Bio-Rad, UK). All PCR amplicons were separated by electrophoresis on a 2% agarose gel prepared by adding 4 g agarose to 200 ml TAE containing 1 μg/ml ethidium bromide and visualized on a UV transilluminator (UVP BioDoc It imaging system, UK). Hyperladder IV and/or Hyperladder I (Bioline, UK) were used as the molecular-weight marker and band positions were determined manually against the marker.
Pulsed-field gel electrophoresis (PFGE)
Chromosomal DNA was prepared by harvesting cells in phosphate-buffered saline and lysed in 1·3% agarose plugs according to the method of Gibson et al. [Reference Gibson, Fitzegerald and Owen32]. The DNA was digested overnight at 25°C using 20 U SmaI (New England Biolabs, UK). PFGE was performed on a DRIII apparatus (Bio-Rad) at 6·6 V/cm for 22 h, with pulse times increasing from 0·5 to 40 s. Standardized parameters were as proposed by CAMPYNET (http://campynet.vetinst.dk). Gels were stained with ethidium bromide (Sigma) and the images captured using UV illumination with a video system. PFGE profiles were recorded using TotalLab TL120-DM (Nonlinear Dynamics, UK). Similarities between profiles were derived using Sorensen's similarity coefficient, which were converted to the distance value E s as described by Sait et al. [Reference Sait33].
Statistical analyses
Samples were grouped by their level of contamination, based on Campylobacter load and competing bacterial flora in different sample types. Samples were grouped by pure faeces (detection hampered by high level of competing flora), high proportion of faeces to sample and low proportion of faeces to sample (detection hampered by low campylobacter numbers). Differences between isolation rates, frequency and number of each species obtained using the three different methods were compared using McNemar's test in GraphPad software (San Diego, USA). For the PFGE data, the differences between the mean values of E s for different subsets within an edited dataset were analysed using Student's t test.
RESULTS
Comparison of three enrichment methods
Campylobacter spp. were isolated from 87%, 73% and 80% of the faecal samples (n=15) using methods based on mExeter, mBolton and Preston broths, respectively. Isolation from this sample type was not improved by using one particular broth over another. For the soil and litter sample group (n=10) Campylobacter spp. were isolated from 100%, 60% and 90% of the samples using mExeter, mBolton and Preston broths, respectively, and none of the enrichment broths offered significantly improved isolation from this sample group.
Campylobacter spp. were isolated from 65%, 53% and 47% of the environmental sample group which included feed, water, bootsock and swabs taken from inside and outside the broiler houses (n=119) using methods based on mExeter, mBolton and Preston broths, respectively. The method using mExeter performed significantly better than Preston (P<0·001), but there was no significant difference in the proportion of positive samples detected between mExeter and mBolton or between mBolton and Preston broths (P>0·05).
Campylobacter spp. detection in relation to enrichment method
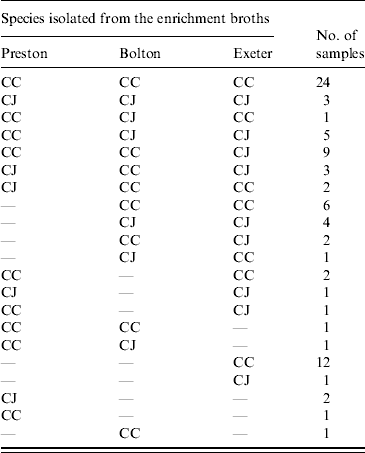
Isolates were confirmed as either C. jejuni or C. coli using a multiplex PCR. For the faecal samples both C. jejuni and C. coli were obtained depending on the enrichment method used. In 8/13 samples there was agreement in the species isolated using at least two enrichment methods (Table 3). There was no significant difference between the three enrichment methods (P>0·05) in the frequency of recovery of each species from faecal samples. Only C. coli was isolated from the soil and litter samples so there was no difference (P>0·05) in the species recovered using the three enrichment methods. Both C. jejuni and C. coli were obtained from the environmental swab, feed and water samples, with both species being obtained from all three enrichment broths. In 61 samples, using at least two enrichment methods, there was agreement in the species isolated (Table 4). However, in 25 samples both C. jejuni and C. coli were isolated depending on the enrichment method used (Table 4). C. jejuni was isolated from significantly more samples using the method based on mExeter compared to Preston (P<0·01) and mBolton (P<0·003) broths but there was no difference between the latter two methods. There was no significant difference between the three enrichment methods for the isolation of C. coli (P>0·05, Table 4). Samples that were only positive in one of the enrichment methods (n=17) were excluded from the analysis as the objective was to compare positivity of individual samples in the different broths. Interestingly, the majority of these (n=13) were obtained using mExeter enrichment broth.
Table 3. Number of faecal samples containing C. jejuni (CJ) and C. coli (CC) in relation to the enrichment protocol used
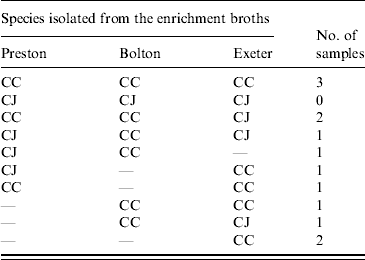
Table 4. Number of environmental (swabs, feed, water) samples containing C. jejuni (CJ) and C. coli (CC) in relation to the enrichment protocol used
Campylobacter subtypes isolated in relation to method
In total, isolates from eight faecal and 25 environmental (swab, feed, water) samples were analysed and 27 different PFGE groups were assigned (Figs 1 and 2). Only samples that were positive in two or more of the enrichment broths were selected for this analysis. Cultures were recovered from frozen storage for the PFGE technique, which starts with a fresh culture. Several isolates despite multiple attempts failed to recover from frozen storage. None of the samples from the soil and litter group recovered so the analysis could not be performed on this sample group. Interestingly, two isolates that were speciated as C. jejuni had an identical PFGE profile to three isolates that were confirmed as C. coli by PCR. Analysis of the average branch lengths as a measure of the diversity of the Campylobacter isolates showed no difference between the three enrichment methods for faecal samples (Fig. 1). However, there was a significant difference between the strains recovered by mExeter and Preston broths (P=0·01) from the environmental samples (Fig. 2). The average branch lengths for Preston and mBolton isolates were shorter than that of mExeter isolates indicating that the former were more clonal. There was no significant difference in the average branch lengths between the strains isolated from mBolton and Preston broths (P=0·10) or between mExeter and mBolton broth (P=0·08). Low numbers of samples were analysed in these groups and it is possible that a lack of statistical significance is due to this rather than a lack of difference between the populations recovered using the different broths.
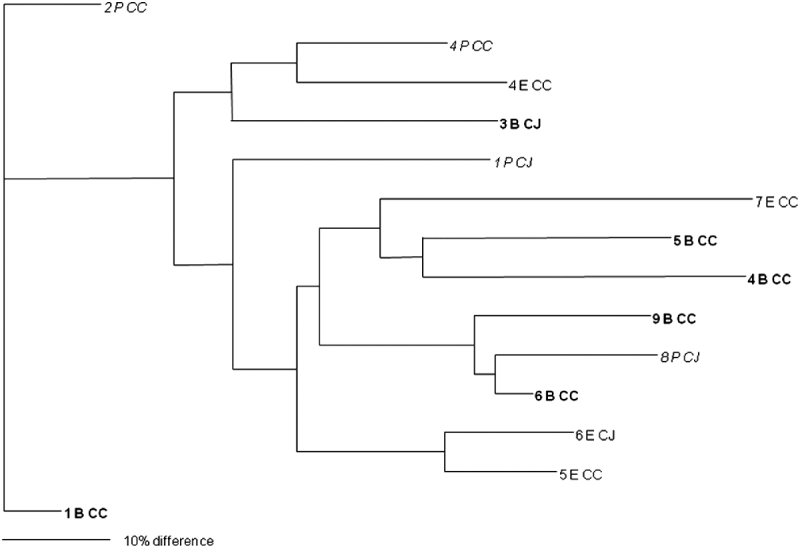
Fig. 1. Dendrogram of relatedness of PFGE profiles of isolates from faecal samples (numbered) enriched in mExeter (E), mBolton (B, in bold) or Preston (P, in italics); C. jejuni (CJ), C. coli (CC). The scale bar represents a 10% difference between isolates. Some isolates could not be recovered from storage and are omitted from this diagram.
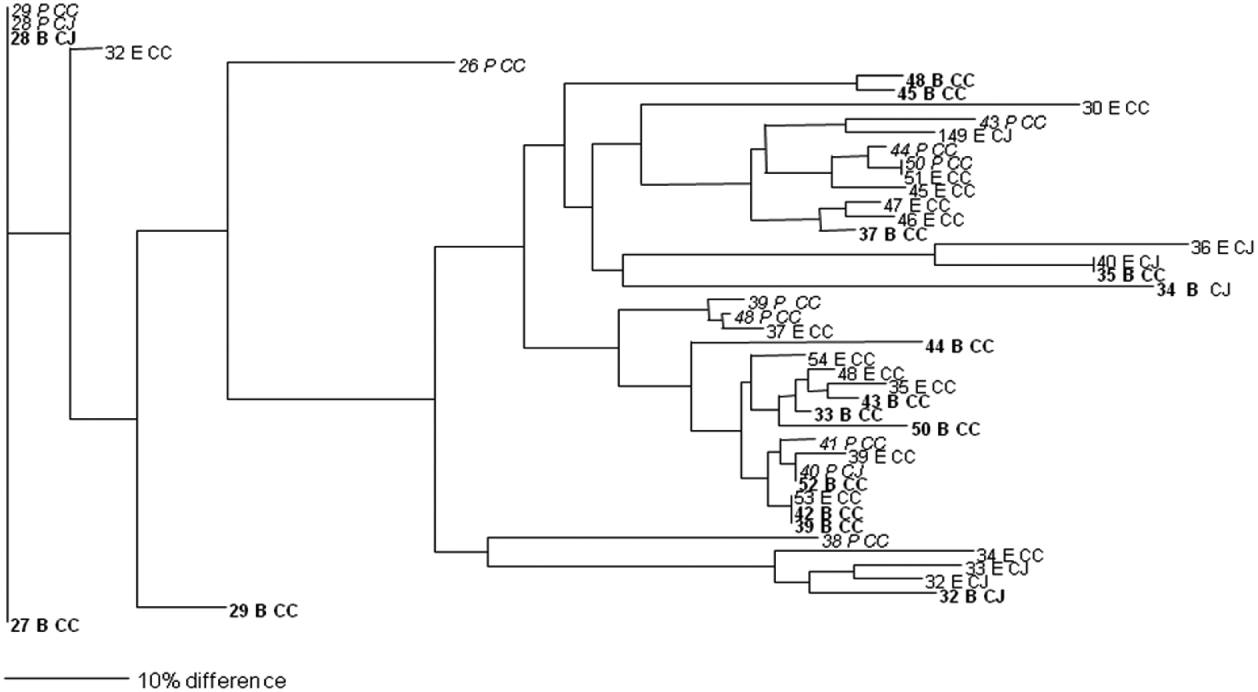
Fig. 2. Dendrogram of relatedness of PFGE profiles of isolates from environmental samples (numbered) enriched in mExeter (E), mBolton (B, in bold) or Preston (P, in italics); C. jejuni (CJ), C. coli (CC). The scale bar represents a 10% difference between isolates. Some isolates could not be recovered from storage and are omitted from this diagram.
DISCUSSION
Enrichment culture is used to recover Campylobacter from samples, such as food, where cells may either be damaged or present in low numbers. Although widely used, there are no standard recommendations [Reference Corry17] and the choice of the enrichment method used varies. Normally, Bolton or mBolton broth, which is endorsed by both the ISO and US FDA, is favoured as it has been shown to be effective in isolating Campylobacter from poultry-related samples [Reference Williams34]. Other enrichments including Preston and mExeter broths have been used successfully to isolate Campylobacter from chicken carcass rinse, environmental, and food samples [Reference Jørgensen5, Reference Josefsen22, Reference Humphrey, Martin and Mason24].
In the first part of this study, the presence of Campylobacter spp. in the farm environment was examined by three enrichment methods. Isolation of these organisms from the swabs of the farm environment, feed and water sample group, where Campylobacter present is likely to be in low numbers, was significantly enhanced using mExeter broth compared to both Preston and Bolton broths. For the faecal sample and the soil and litter sample groups, where the numbers of Campylobacter spp. present is likely to be higher, the three enrichment methods performed equally, mExeter broth appeared to offer improved isolation from these samples but due to the small sample size the differences were not significant. Other studies have shown that other enrichment methods offered improved isolation of Campylobacter spp. from environmental and food samples compared to Preston broth [Reference Humphrey, Martin and Mason24, Reference Paulsen25, Reference Williams34]. Baylis et al. [Reference Baylis28] found that Bolton broth offered improved isolation compared to Preston broth for the detection of Campylobacter spp. in naturally contaminated samples. Moreover, Exeter broth showed marked superiority over Preston and Park & Sanders broths for the investigation of contaminated chicken meat and water samples [Reference Humphrey, Martin and Mason24]. We are not aware of any previous publications comparing the sensitivity of Exeter or mExeter and Bolton or mBolton enrichment broths.
All three broths could support the growth of both species. Some broths, e.g. mExeter, were better for the isolation of C. jejuni than the others; C. coli isolation was similar in all broths. Both C. coli and C. jejuni were detected in the environmental swabs taken, feed, water and faecal samples from the two farms sampled but only C. coli was isolated from the soil and litter samples. The presence of both species in the environment and chicken has been found in other studies [Reference Humphrey35].
Campylobacter isolated from all samples were characterized using a multiplex PCR developed in this study that combined primers for the hipO and lpxA genes for C. jejuni with the glyA and lpxA genes for C. coli; the hipO and glyA genes have been used successfully in combination in a previous study [Reference Wang36]. Similarly, the lpxA primers have been used previously with other primers designed for identification of C. lari and C. upsaliensis [Reference Klena29]. Further characterization using PFGE found that for the majority of isolates of the same species from both the faecal and environmental samples were genotypically identical or closely related. However, isolates recovered from environmental (swabs, feed, water) samples, using Preston broth were less genetically diverse, and appeared more clonal than those recovered with mExeter. This broth recovered more C. jejuni compared to the other two methods and this could be a possible explanation for the greater diversity seen when using the mExeter method. On 18 occasions, a different PFGE type was identified in the different enrichment broths from the same sample. This result could be due to the selective components of the broth favouring a particular isolate or a broth-induced stress altering the isolate recovered; this observation merits further investigation. PFGE was performed on a single colony from each of the three enrichment broths used on an individual sample. It is possible that strains of Campylobacter spp. other than those isolated could have been present in the original sample but not detected from a single colony pick. We therefore cannot say whether the method used selects for a single strain, or biases the population towards certain strains. However, examination of all of the isolates from each enrichment broth across all samples shows that enrichment does bias the population obtained, but the dynamics of this on the level of a single enrichment would require further work using multiple colony selections from each enriched sample.
The identical PFGE pattern seen in isolates identified as C. coli and C. jejuni by multiplex hipO/glyA/lpxA PCR raises the possibility that these strains may be more closely related than the results suggest. PCR tests for single genes revealed that these strains carried genes normally associated with both C. coli and C. jejuni (data not shown). Sheppard et al. [Reference Sheppard37] have suggested that these C. jejuni and one clade of C. coli are undergoing continual recombination and despeciation in the farm environment and that hybrid strains containing genes from each species exist there. This mechanism is a possible explanation for the results seen in this work, as it explains why some strains could test positive for both C. coli and C. jejuni genes. Whole genome sequencing, and identification of the clades to which the Campylobacter strains identified here belong are required to confirm this hypothesis.
In conclusion, the enrichment methods used have an important role in terms of detection from naturally contaminated samples affecting both the number of positive samples obtained and species of Campylobacter isolated. The use of Preston broth resulted in more false-negative results and applied a greater population bias compared to mExeter but not mBolton broths. The fact that both C. jejuni and C. coli were present in a considerable proportion of samples, but that individual enrichment methods biased recovery towards one species, indicates that the enrichment method used influences the species recovered.
ACKNOWLEDGEMENTS
The sample collection and isolation part of this study was funded by Defra (project OZ0608). We thank Dr Nicola Elviss for helpful discussions in the initial stages of this work and Dr Victoria Morris for help with PFGE. We thank the farmers who allowed us access to their farms to collect samples and we acknowledge Debra Langton for excellent technical assistance.
DECLARATION OF INTEREST
None.








