Colorectal cancer (CRC) is the third most malignant tumour worldwide, accounting for approximately 10 % of all cancer incidence and mortality(Reference Jemal, Bray and Center1, Reference Jemal, Siegel and Xu2). Despite the considerable effort dedicated to CRC diagnosis and therapeutic strategy over the past two decades, one-fifth of the patients are still being diagnosed with CRC in the advanced stage, which is usually treated by a combination of surgery and adjuvant chemotherapy(Reference Lin, Gray and Jemal3–Reference Zauber, Winawer and O’Brien5). Most patients are resistant to chemotherapy, and the cancer may occur again in these patients with eventual metastasis(Reference Pessaux, Chenard and Bachellier6). A number of recent studies have reported significant association of microRNA (miRNA) expression with the progression of CRC(Reference Srivastava and Srivastava7–Reference Bhardwaj, Singh and Singh9). Therefore, based on RNA network analysis, it would be desirable to search for a novel targets that can be used in the prevention or therapy of CRC.
miRNA are endogenous small non-coding RNA with 18–22 nt in length, and they regulate gene expression by interfering with the translation of mRNA through binding to their 3′-UTR (untranslated regions), directly degrading the bound mRNA(Reference Bartel10, Reference Cho11). Aberrant expression of miRNA can give rise to either tumour suppressors or oncogenes in many human cancers. In CRC, several miRNA have been identified as essential for tumour development. The differential expression of miRNA in human CRC tissues and normal colorectal mucosa was first reported by Michael et al. (Reference Michael, O’Connor and van Holst Pellekaan12), and twenty-eight differentially expressed miRNA were detected, including miR-320, miR-321, miR-200c, miR-223 and miR-145. Monzo et al. (Reference Monzo, Navarro and Bandres13) found that miR-17-5p can target E2F transcription factor 1 present in the early embryonic development and tumourigenesis of human colon, regulating cell proliferation in both cases. Other investigators have reported that miR-34a directly and negatively controls a series of downstream target genes of p53, forming a miR-34a-p53 positive feedback loop(Reference Tazawa, Tsuchiya and Izumiya14). Bandres et al. (Reference Bandres, Bitarte and Arias15) found that miRNA-451 can regulate the expression of migration inhibitory factor (MIF), thereby, inhibiting the proliferation of CRC cells and enhancing the sensitivity of the tumour to radiotherapy. In addition, miRNA-320a and miR-139 can target β-catenin and RAP1B (member of the RAS oncogene family), respectively, to inhibit the proliferation of human colon cancer cells(Reference Guo, Hu and Ge16, Reference Sun, Huang and Li17). miR-24 was first discovered by Lagos-Quintana et al. (Reference Lagos-Quintana, Rauhut and Lendeckel18) in 2001 when non-spine animals and spine animals were studied. The human miR-24 consists of miR-24-1 and miR-24-2, and are located on chromosomes 9 and 19, respectively, and the mature sequences include miR-24-3p, miR-24-1-5p and miR-24-2-5p(Reference Sun, Zhang and Yang19). Aberrant expression of miR-24-1 in several types of cancer tissues has been reported(Reference Guo, Fu and Chen20–Reference Qin, Shi and Zhao26). However, the functional role of miR-24-1-5p in CRC is still unknown.
Black raspberry (BRB) belongs to the Rubus occidentalis families of berries, and they are rich in flavonoids and phenolic acids(Reference Gu, Ahn-Jarvis and Riedl27, Reference Wang, Hecht and Carmella28). The chemopreventive effect of BRB against CRC has been well documented by studies carried out by our research group and those of others(Reference Montrose, Horelik and Madigan29–Reference Bi, Fang and Wang32). However, few studies have focussed on the role of miRNA in association with the chemopreventive activity of BRB in CRC and the underlying mechanisms.
In this study, we hypothesised that miR-24-1-5p might play an important role in the initiation and progression of CRC, and that it might also be involved in the chemopreventive effect of BRB against CRC. To test this hypothesis, we conducted one in vivo study with mice and several in vitro studies with human cell lines, and found that miR-24-1-5p could function as a tumour suppressor by targeting several β-catenin-related oncogenes involved in specific cancer-related pathways in CRC. Our current finding might facilitate the development of novel diagnostic and therapeutic strategies for the prevention and treatment of CRC.
Methods
Chemicals and reagents
Azoxymethane (AOM, A5486) was purchased from Sigma-Aldrich Inc. Dextran sulphate sodium (DSS, 0216011091) was purchased from MP Inc. Jianfeng Nature Products Technology Co., Ltd provided the BRB anthocyanins (purity >90 %). BRB anthocyanins consist of three major anthocyanins cyanidin-glucoside, cyanidin-xylosylutinoside and cyanidin-rutinoside, and the contents are 2·63, 0·73 and 16·91 mg/g, respectively. The cyanidin-sambubioside is less abundant, the detail of which can be found in our recently published paper(Reference Xiao, Guo and Bi33). β-Catenin, cyclin D1, c-Myc and cyclin-dependent kinase 4 (CDK4) were obtained from Cell Signaling Technology Inc. β-Actin, E-cadherin, glycogen synthase kinase 3 β (GSK3-β), phospho-glycogen synthase kinase 3 β (p-GSK3-β), B-cell lymphoma-2 (BCL-2) and haemagglutinin were obtained from Sangon Biotech Co., Ltd. Secreted frizzled-related protein 2 (SFRP2) was bought from Abcam Inc. Secreted frizzled-related protein 5 (SFRP5) was obtained from Biorbyt Explore Bioreagents. Goat anti-rabbit and goat anti-mouse IgG conjugated with horseradish peroxidase used as secondary antibodies in Western blot were obtained from ComWin Biotech Co., Ltd.
Animal care and experimental protocol
A total of 5-weeks-old male C57BL/6J mice (18–20 g, Benxi Changsheng Laboratory Animal Technology Co., Ltd) were given water ad libitum, and maintained under a 12 h light–12 h dark cycle. The animals were housed in cages (five animals per cage) in a room with temperature set at 21 ± 2·0°C and humidity controlled at 50 ± 5 %. All animal experiment protocols were approved by the Ethics Committee of Liaoning University of Traditional Chinese Medicine (Shenyang, China).
Establishment of colitis-induced colorectal cancer mice model and black raspberry anthocyanins supplementation
After 1 week of adaptation, the mice were given AOM (10 mg/kg body weight) in a single intraperitoneal injection. In all, 1 week later, they were given 2 % DSS in the drinking water for 1 week, followed by normal water for another 2 weeks, and this cycle was repeated two more times.
The mice were divided into three groups: one healthy control group and two AOM/DSS-treated groups. Each group consisted of ten animals. Mice in the healthy control group were not given any treatment, and they were fed with a chow diet over a 9-week period. As for the two AOM/DSS-treated groups, one group was fed with just the chow diet, whereas the other group was fed with the chow diet containing 7·0 µmol/g BRB anthocyanins (10% BRB). The concentration of BRB anthocyanins in the diet corresponded to the anthocyanins content in a 10 % freeze-dried BRB powder, and it was chosen on the basis of previous study(Reference Bi, Fang and Wang32, Reference Stoner, Wang and Zikri34). The formula composition of the chow diet and BRB anthocyanins-supplemented diet is listed in Table 1. The diets were stored at –20°C before they were used in the experiment.
Table 1 Composition of chow diet and black raspberry (BRB) anthocyanins-supplemented diet

At the end of 9th week, the mice were sacrificed by CO2 inhalation followed by cervical dislocation. The colon tissue was immediately frozen in liquid N2, and then stored at –80°C for miRNA array analysis.
MicroRNA microarray expression profiling and data analysis
miRNA expression profiles were generated using the Agilent Mouse miRNA (8 × 60K, design ID: 070155; Agilent Technologies Inc.). Total RNA was quantified by the NanoDrop ND-2000 (Thermo Scientific). RNA integrity was assessed by an Agilent Bioanalyzer 2100 (Agilent Technologies Inc.). Total RNA was dephosphorylated, denaturated and then labelled with cyanine-3-cytidine triphosphate. After purification, the labelled RNA was hybridised to the microarray. After washing, the array was scanned with an Agilent Scanner G2505C (Agilent Technologies Inc.). Image of the array was acquired by Feature Extraction software (version 10.7.1.1; Agilent Technologies Inc.). Genespring software (version 13.1; Agilent Technologies Inc.) was used to analyse the raw data.
RNA extraction and quantitative real-time PCR
Total RNA was isolated from the cells using a TRIzol Reagent Kit (ComWin Biotech Co., Ltd), and the first-strand complementary DNA (cDNA) encoding the miRNA with poly A was generated by reverse transcription using a miRNA cDNA synthesis kit (ComWin Biotech Co., Ltd). Real-time PCR (RT-PCR) was performed using SYBR Green assay with specific primers. The assay was conducted with a miRNA real-time assay kit (ComWin Biotech Co., Ltd) and amplification was performed on an Applied Biosystems 7500 real-time PCR system in accordance with the manufacturer’s instructions. The sequences of the primers were: mmu-miR-24-1-5p: forward 5′-AGTGCCTACT GAGCTGATATCAGT-3′; hsa-miR-24-1-5p: forward 5′-GTGCCT ACTGAGCTGATATCAGT-3′; β-catenin: forward 5′-GCTGGTG ACAGGGAAGACAT-3′, reverse 5′-AGGCAAACCGTGAAAA GATG-3′; cyclin D1: forward 5′-AACTACCTGGACCGCTTCCT-3′, reverse 5′-CCACTTGAGCTTGTTCACCA-3′; c-Myc: forward 5′-TTCGGGTAGTGGAAAACCAG-3′, reverse 5′-CAGCAGCTCG AATTTCTTCC-3′; CDK4: forward 5′-ATTGGTGTCGGTGCCT ATG-3′, reverse 5′-AACTGTGCTGATGGGAAGG-3′. The expression levels of these genes were normalised to that of the internal control (U6: forward 5′-ATTGGAACGATACAGAG AAGATT-3′, reverse 5′-GGAACGCTTCACGAATTTG-3′ or β-actin: forward 5′-AGGCAAACCGTGAAAAGATG-3′, reverse 5′-AGGCAAACCGTGAAAAGATG-3′) according to the 2–ΔΔC T method.
Plasmid constructs and transfection
The mature sequence of miR-24-1-5p is UGCCUACUGAGCUGAUAUCAGU. The sequence of human pri miR-24-1-5p was constructed using the following primers: miR-24-1-5p forward, 5′-CCGCTCGAGCAACAGGGTTTTCCAAGTCTAC-3′ and miR-24-1-5p reverse, 5′-GGAAGATCTTCACCTAAGTCGGAAAT CATGTGGTA-3′. The PCR product was digested with XhoI and BglII and then inserted into XhoI-BglII cut pcDNA3-haemagglutinin. HCT-116 and Caco-2 cells were then transfected with the human pri-miR-24-1-5p using lipofectamine 2000 (Invitrogen). Cells transfected with the blank plasmid were used as negative controls. The constructed human pri-miR-24-1-5p was validated by sequencing.
Cell culture
Human CRC cell lines HCT-116 (CCL-247; American Type Culture Collection) and Caco-2 (Cell Bank of Beijing Cancer Institute) were cultured in Dulbecco’s modified Eagle’s medium and α-minimum essential medium (Hyclone, Thermo Fisher Scientific Inc.), respectively. In both cases, the medium was supplemented with 10 % (v/v) fetal bovine serum (Dalian Meilun Biology), 10 000 U/ml penicillin G, 10 mg/ml streptomycin, and the cultures were incubated in a 5 % CO2 humidified atmosphere at 37°C.
Wound healing assay
HCT-116 and Caco-2 cells with 70 % confluency were used in the wound healing assay. First, a sterile pipette tip was used to make a straight-line scratch in the cell monolayer. The cells were washed with PBS followed by the addition of fresh medium. They were then transfected with miR-24-1-5p, and after 24 h, the medium was discarded, and the gap remaining in the scratch was calculated by photomicrograph.
Cell proliferation and colony formation assays
Cell viability was assessed by the 3-(4, 5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay. Briefly, 24 h after transfection with miR-24-1-5p, the Caco-2 and HCT-116 cells were incubated for 4 h with 0·5 mg/ml MTT reagent. The culture medium was then removed, and 100 µl dimethyl sulfoxide was added to each well to dissolve the formazan crystals. The absorbance of the plate was measured at 490 nm. To further investigate the role of miR-24-1-5p in cancer cell survival, the cells were seeded in six-well plates at 200/well and grown for 2 weeks for colony formation assay. The colonies of cells that appeared were first washed with PBS, and then fixed with methanol followed by staining with 0·5 % Giemsa. The number of colonies appearing in each well was counted using an inverted microscope.
Transwell migration assay
Caco-2 and HCT-116 cells were seeded on the top chamber of a transwell, and serum-free medium was added to the bottom chamber. After 24 h, the cells on top of the membrane were removed with a cotton swab, and the membranes of the transwell inserts were then fixed with 4 % paraformaldehyde. The cells that migrated to the bottom of the membrane were stained with 4′,6-diamidino-2-phenylindole (Beijing Dingguo Changsheng Biotechnology Co., Ltd). A total of ten fields were randomly selected from each insert and captured under a microscope at 20× magnification. Cell numbers were quantified using Image-Pro Plus 6.0. To calculate the migration index, the number of cells obtained for each treated group was normalised to the number of cells in the control group.
Target prediction of microRNA
Target genes of miR-24-1-5p were predicted using algorithms, MiRanda version 3.3a, with the default parameters and cutoffs (score S≥140 and energy E≤–7·0) and website tools (http://www.microrna.org/microrna/home.do).
Western blotting
Cells were lysed in chilled radioimmunoprecipitation assay buffer containing 1 mmol/l phenylmethylsulfonyl fluoride (Beijing Dingguo Changsheng Biotechnology Co., Ltd) for 30 min on ice. After centrifugation at 14 000 g/min for 15 min at 4°C, the supernatant was collected and a sample of the supernatant containing 40 µg protein was resolved by SDS–PAGE using 10 % gel. After electrophoresis, the proteins in the gel were transferred to a nitrocellulose membrane (Sangon Biotech Co., Ltd), and the membrane was blocked with 5 % bovine serum albumin fraction V (Roche) in Tris-buffered saline and Tween 20 buffer for 2 h. It was then incubated with the appropriate primary antibody for 3 h at room temperature followed by the corresponding secondary antibody. Signals on the blot were detected by an enhanced chemiluminescence technique (Amersham Life Science).
Immunohistochemical staining
Mouse intestinal tissues from AOM/DSS-induced mice with or without BRB anthocyanins were collected, fixed in 10 % buffered formalin and embedded in paraffin as previously reported(Reference Bi, Tong and Dockendorff35). Sections (4 µm thick) of the tissue were used for immunohistochemical staining using anti-E-cadherin antibody (Santa Cruz Biotechnologies). The procedure was similar to that previously reported(Reference Bi, Tong and Dockendorff35). In brief, the sections were deparaffined, rehydrated, quenched with 3 % H2O2, blocked with 10 % normal serum and then incubated with β-catenin antibody for overnight at 4°C followed by incubation with secondary antibody and avidin–biotin complex (ABC kit; Vector Laboratory). The staining was developed using 3′,5′-diaminobenzidine followed by lightly counter-staining with haematoxylin. Image of the stained slide was captured by the Aperio Image system.
Statistical methods
The statistical software SPSS 19.0 was used to analyse the data. To compare differences within and between groups, t tests and ANOVA followed by Fisher’s least significant difference analysis were used. Statistical significance was considered at the P <0·05 level.
Results
Tissue microRNA microarray profiling
To elucidate the miRNA-mediated tumourgenic pathways in CRC, identifying the aberrantly expressed miRNA would be the first step. Based on this point, we performed tissue miRNA microarray analysis to obtain the global miRNA expression signatures in the colon tissue of AOM/DSS-induced mice with or without BRB anthocyanins supplementation. Comparison of the results from the two groups showed that several miRNA were differentially regulated by the administration of BRB anthocyanins (Fig. 1(a)). The result obtained from the hierarchical clustering analyses of the tissue arrays is presented in Fig. 1(b). Low level of miR-24-1-5p was expressed in the AOM/DSS-induced group, but after 9 weeks of feeding with BRB anthocyanins, the level of mir-24-1-5p was significantly up-regulated, suggesting that miR-24-1-5p might have a suppressive effect on tumour formation. This also confirmed the chemopreventive property of BRB anthocyanins.
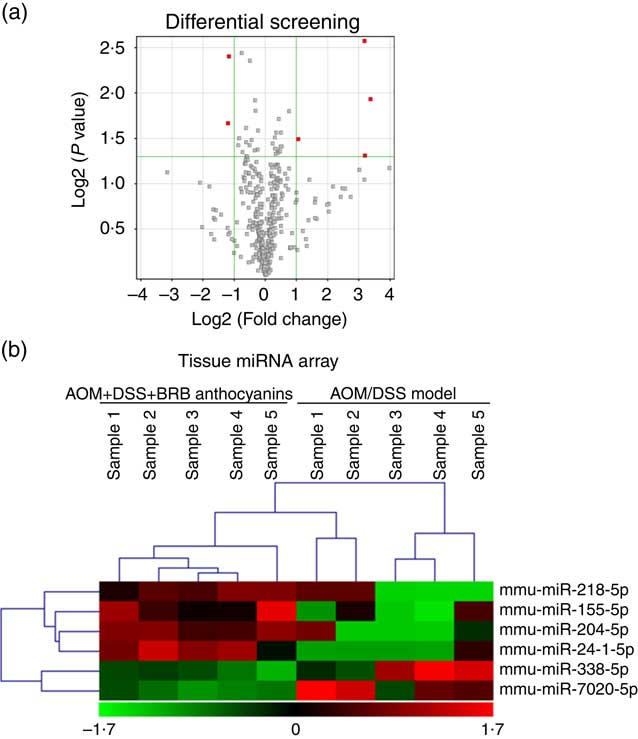
Fig. 1 Differential expression of microRNA (miRNA) in mice colon tissue. (a) Up-regulated miRNA are indicated in red. (b) Tissue miRNA array. Green – down-regulation; red – up-regulation (n 5). AOM, azoxymethane; DSS, dextran sulphate sodium; BRB, black raspberry.
Expression of miR-24-1-5p is down-regulated in the colon tissue of azoxymethane/dextran sulphate sodium-induced mice and up-regulated in human colorectal cancer cells lines treated with black raspberry anthocyanins
To further evaluate the result shown by the microarray analysis using mouse model, miR-24-1-5p expression in the colon tissue of the AOM/DSS-induced group with or without BRB anthocyanins administration was analysed by quantitative RT-PCR (qRT-PCR). The expression pattern of miR-24-1-5p obtained by qRT-PCR was consistent with the microarray data (Fig. 2(a)). The expression of miR-24-1-5p was also determined in several human CRC cell lines (Caco-2, HCT-116, Lovo, HT29 and SW480) after treatment with different doses of BRB anthocyanins. BRB anthocyanins caused the level of miR-24-1-5p to increase. The pattern of miR-24-1-5p expression in BRB anthocyanins-treated human cancer cells was consistent with the miRNA microarray and qRT-PCR data obtained for the colon tissues of mice fed with the BRB anthocyanins-supplemented diet (Fig. 2(b)).

Fig. 2 miR-24-1-5p expression in colon tissue and colorectal cancer (CRC) cells as detected by quantitative RT-PCR (qRT-PCR). (a) qRT-PCR analysis of miR-24-1-5p levels in colon tissue of azoxymethane (AOM)/dextran sulphate sodium (DSS)-induced mice with or without black raspberry (BRB) anthocyanins supplement. (b) Relative miR-24-1-5p expression in different CRC cell lines. One-way ANOVA was used to compare different groups of human CRC cell lines or mouse colon tissues. Values are means (n 5), with their standard errors represented by vertical bars. Mean values were significantly different when compared with the solvent control: * P <0·05, ** P <0·01, *** P <0·001. (a) ![]() , Control;
, Control; ![]() , AOM+DSS model;
, AOM+DSS model; ![]() , AOM+DSS+BRB anthocyanins. (b)
, AOM+DSS+BRB anthocyanins. (b) ![]() , Control;
, Control; ![]() , 25 μg/ml BRB anthocyanins;
, 25 μg/ml BRB anthocyanins; ![]() , 50 μg/ml BRB anthocyanins.
, 50 μg/ml BRB anthocyanins.
β-Catenin in azoxymethane/dextran sulphate sodium-induced mice is reduced by black raspberry anthocyanins
To determine the effect of β-catenin in AOM/DSS-induced mice regulated by BRB anthocyanins in situ, we performed immunohistochemical staining. AOM/DSS-induced mice that were given the BRB anthocyanins-supplemented diet showed significantly reduced levels of β-catenin in the small intestine compared with AOM/DSS-induced mice given the BRB anthocyanins-free diet (Fig. 3).
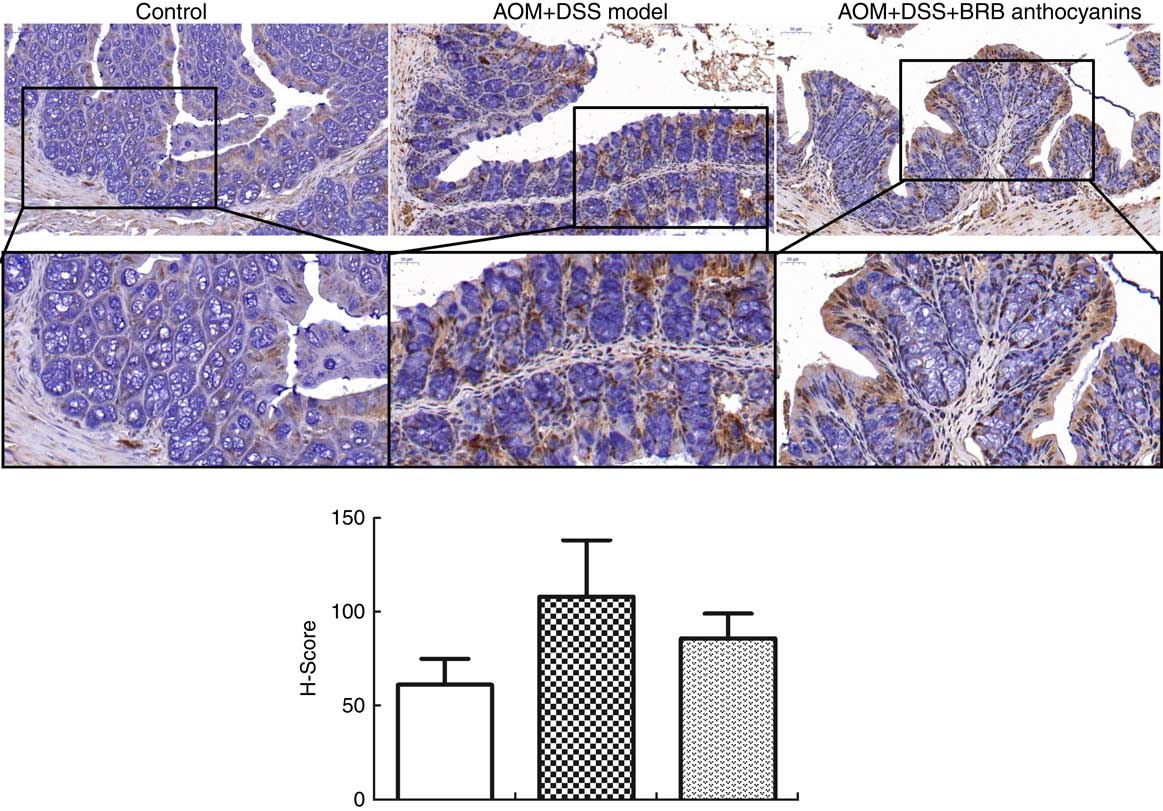
Fig. 3 Effect of black raspberry (BRB) anthocyanins supplement on β-catenin expression in mouse colon tissue. In situ β-catenin expression in the intestinal epithelial cells of azoxymethane (AOM)/dextran sulphate sodium (DSS)-induced mice with or without BRB anthocyanins supplement as detected by immnunohistochemical staining (anti-β-catenin antibody dilution 1:100) (magnification 20 × and 40 ×, respectively). ![]() , Control;
, Control; ![]() , AOM+DSS model;
, AOM+DSS model; ![]() , AOM+DSS+BRB anthocyanins.
, AOM+DSS+BRB anthocyanins.
Effects of restoring miR-24-1-5p expression on colorectal cancer cell proliferation and migration
To further evaluate the biological function of miR-24-1-5p, human HCT-116 and Caco-2 cells were transfected with miR-24-1-5p and the viability of the cells was determined by MTT assay. Both miR-24-1-5p-transfected HCT-116 and Caco-2 cells exhibited markedly reduced cell viability (56 % in the case of HCT-116 and 52 % for Caco-2 cells) compared with the corresponding control cells (Fig. 4(a) and (b)). These cells also displayed significantly reduced sclerosis as revealed by wound healing assay (Fig. 4(c) and (d)). Furthermore, they exhibited significant reduction in cell migration and cell survival as revealed by transwell assay (Fig. 4(e) and (f)) and colony formation assay (Fig. 4(g) and (h)), respectively. Taken together, the results suggested that overexpression of miR-24-1-5p could significantly decrease the proliferation, migration and survival of these human CRC cell lines, thereby indicating that miR-24-1-5p might play a tumour-suppressive role in CRC by reducing or suppressing the growth of the tumour cells.
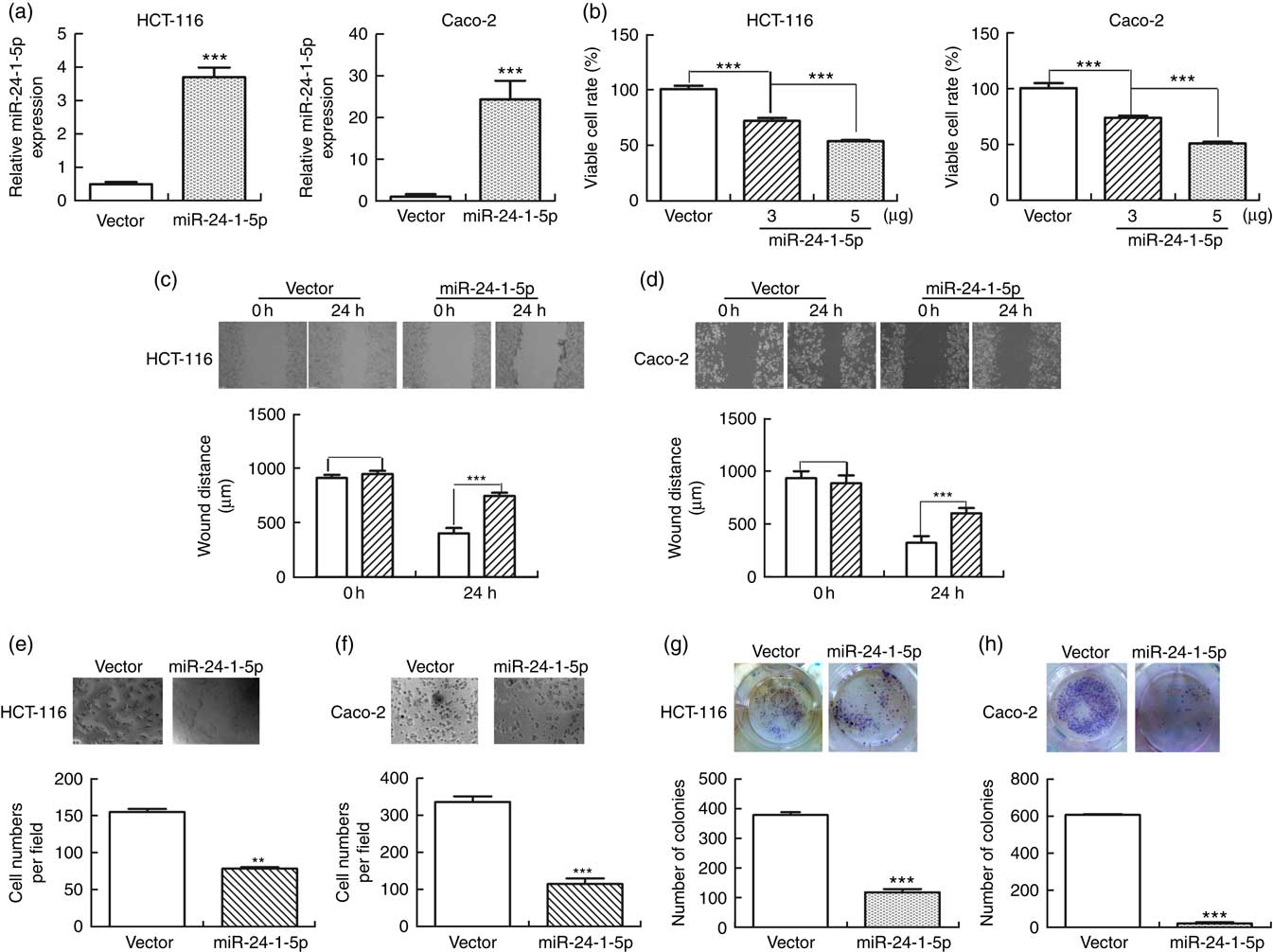
Fig. 4 miR-24-1-5p inhibits human colorectal cancer cells proliferation, migration and formation. HCT-116 and Caco-2 cells were transfected with 5 µg miR-24-1-5p for 24 h and then subjected to the following analyses: (a) expression level of miR-24-1-5p was measured by quantitative RT-PCR. (b) Cell viability was determined by 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide assay. (c, d) Cell migration was evaluated by wound healing assay. The inhibitory effect on cell migration was determined by an increase in the percentage of the wound area remaining at 24 h. Wound area at 0 h was taken as 100 %. ![]() , Vector;
, Vector; ![]() , miR-24-1-5p. (e, f) Cell invasion was examined by microscopy. (g, h) Representative micrographs and quantitative analysis of colony formation. Differences within and between groups were compared by one-way ANOVA followed by Fisher’s least significant difference analysis. Values are means (n 3), with their standard errors represented by vertical bars. Mean values were significantly different when compared with vector control: ** P <0·01, *** P <0·001.
, miR-24-1-5p. (e, f) Cell invasion was examined by microscopy. (g, h) Representative micrographs and quantitative analysis of colony formation. Differences within and between groups were compared by one-way ANOVA followed by Fisher’s least significant difference analysis. Values are means (n 3), with their standard errors represented by vertical bars. Mean values were significantly different when compared with vector control: ** P <0·01, *** P <0·001.
miR-24-1-5p regulates β-catenin
The target genes that miR-24-1-5p may bind to were predicted using the bioinformatics method, miRanda version 3.3a, with default parameters and cutoffs (score S≥140 and energy E≤–7·0). β-Catenin was selected as a potential target among all of the candidates after considering the important role that β-catenin plays in CRC development. A binding domain was detected at the 3′-UTR of β-catenin where it might interact with miR-24-1-5p (Fig. 5(a)). Furthermore, to investigate the influence of miR-24-1-5p on β-catenin expression, miR-24-1-5p was overexpressed in human HCT-116 and Caco-2 cells, and the changes in β-catenin level were measured by qRT-PCR and Western blot. In both cell lines, the protein level of β-catenin was significantly decreased whereas its mRNA level remained unchanged when miR-24-1-5p was overexpressed (Fig. 5(b) and (c)).

Fig. 5 miR-24-1-5p suppresses β-catenin expression via direct binding to the 3′-UTR (untranslated regions). (a) Sequence alignment of putative miR-24-1-5p binding sites in β-catenin 3′-UTR. (b) Western blot analysis of β-catenin protein level in HCT-116 and Caco-2 cells after being transfected with miR-24-1-5p. (c) Quantitative RT-PCR analysis of β-catenin mRNA level in HCT-116 and Caco-2 cells after being transfected with miR-24-1-5p. (d) β-Catenin protein level after treatment with 10 µm cycloheximide (CHX) for different times (0, 10, 12, 14 h). Differences within and between groups were compared by one-way ANOVA followed by Fisher’s least significant difference analysis. ![]() , Vector;
, Vector; ![]() , miR-24-1-5p. HA, haemagglutinin.
, miR-24-1-5p. HA, haemagglutinin.
Further experiment was carried out in which HCT-116 and Caco-2 cells were transfected with miR-24-1-5p or the control vector for 24 h, and cycloheximide (10 µm) was then added to the cells for different time intervals before the cells were subjected to Western blot analysis to determine the protein synthesis rate of β-catenin. The result showed that the protein level of β-catenin in the cells transfected miR-24-1-5p plasmid was significantly decreased compared with the β-catenin level in the cells transfected with the control plasmid, indicating that miR-24-1-5p could promote the degradation of β-catenin protein to influence its stability. The result also suggested that miR-24-1-5p might negatively regulate β-catenin by directly binding to the 3′-UTR of the β-catenin mRNA.
Genes associated with the wnt/β-catenin pathway are regulated by miR-24-1-5p
The impact of miR-24-1-5p over-expression on several downstream target genes of the β-catenin signalling pathway was also investigated. Changes in cyclin D1, c-Myc and CDK4 expression levels in HCT-116 and Caco-2 cells were determined using Western blot and qRT-PCR. Both the mRNA and protein levels of these genes in HCT-116 and Caco-2 cells decreased 24 h after they were transfected with miR-24-1-5p (Fig. 6(a) and (b)). Furthermore, other genes related to the Wnt/β-catenin pathway such as SFRP2, SFRP5, E-cadherin, GSK3-β, p-GSK3-β, and BCL-2 were also analysed. The expression of E-cadherin, p-GSK3-β and SFRP5 was up-regulated, whereas the expression of BCL-2 was down-regulated, while no significant changes in expression were observed for GSK3-β and SFRP2 (Fig. 6(c)). In summary, the data from in vitro human cancer cells demonstrated that β-catenin might be a direct target of miR-24-1-5p, mediating the effect of miR-24-1-5p by affecting the expression of its downstream target genes.
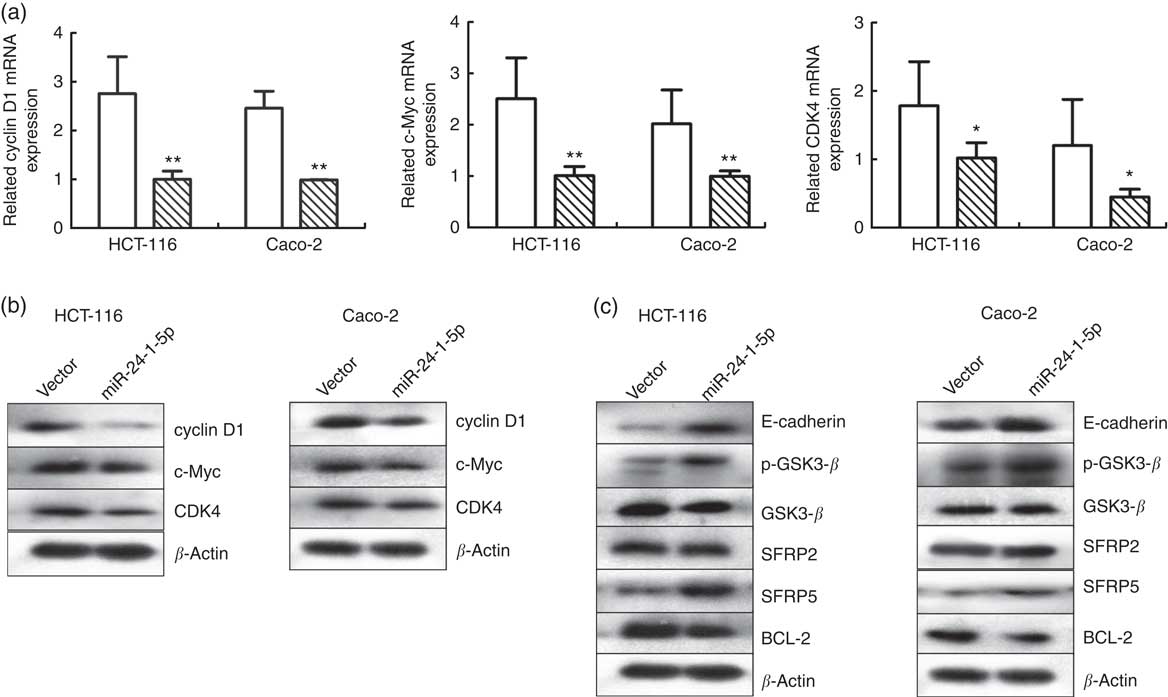
Fig. 6 Effects of miR-24-1-5p on genes associated with Wnt/β-catenin signalling in colorectal cancer cells. Quantitative RT-PCR (a) or Western blot (b) analysis of cyclin D1, c-Myc and cyclin-dependent kinase 4 (CDK4) mRNA in HCT-116 and Caco-2 cells after transfection with 5 µg miR-24-1-5p 24 h (c) Western blot analysis of glycogen synthase kinase 3 β (GSK3-β), secreted frizzled related protein 2 (SFRP2), secreted frizzled related protein 5 (SFRP5), B-cell lymphoma-2 (BCL-2), E-cadherin and phospho-glycogen synthase kinase 3 β (p-GSK3-β) protein levels in HCT-116 and Caco-2 cells 24 h after transfection with miR-24-1-5p. Differences within and between groups were compared by one-way ANOVA followed by Fisher’s least significant difference analysis. Mean values were significantly different when compared with the vector control: * P <0·05, ** P <0·01. ![]() , Vector;
, Vector; ![]() , miR-24-1-5p.
, miR-24-1-5p.
Discussion
Accumulated evidence suggests that the tightly regulated RNA network in a cell can be disrupted by aberrantly expressed miRNA, consequently triggering cancer development and metastasis. The importance of miRNA in various human cancers suggests that modulation of miRNA expression could be a novel strategy for cancer chemoprevention and therapy(Reference Xuan, Yang and Zhao36). In the present study, miR-24-1-5p was significantly up-regulated in AOM/DSS-induced mice fed a diet containing BRB anthocyanins. Furthermore, mechanistic study indicated that miR-24-1-5p might play a tumour-suppressive role, and this was supported by its ability to inhibit cell proliferation, migration and survival, as well as colony formation when it was overexpressed in human CRC cells (Fig. 3). The tumour-suppressive role of miR-24-1-5p appeared to be mediated by its effect on the β-catenin signalling pathway (Figs. 4 and 5).
Implication of the Wnt/β-catenin signalling pathway in cancer has been demonstrated by the effects of this signalling pathway on the growth, metabolism, angiogenesis, migration and metastasis of CRC cells(Reference Zhang, Zhu and Wu37). Aberrant activation of β-catenin and its downstream target genes, which include c-Myc, cyclin D1 and CDK4, can accelerate the initiation and progression of CRC. Our result clearly showed that in human CRC cells, these genes could be regulated through altering the expression of miR-24-1-5p. Indeed, over-expression of miR-24-1-5p in HCT-116 and Caco-2 cells down-regulated the expression of β-catenin, c-Myc, cyclin D1 and CDK4 (Fig. 6). Elucidating the cancer pathways and targets regulated by miR-24-1-5p via β-catenin would provide new insight into the process of oncogenesis for human CRC. Unfortunately, due to ethical issues we could not obtain direct evidence from human subjects to support our claim. However, further research is necessary to characterise the feasibility of targeting miR-24-1-5p in CRC as a cost-effective and simple therapeutic strategy for combating CRC at the clinical level. Much research has been devoted to BRB anthocyanins to investigate its usage as a chemoprevention agent for CRC. This aspect has been widely investigated in animal models and clinical patients. The mechanism underlying the chemopreventive activity of BRB anthocyanins involves induction of cell apoptosis and differentiation, and reduced cell proliferation, inflammation, angiogenesis and invasiveness(Reference Li, Zhang and Stoner38). The chemopreventive effect of BRB anthocyanins on CRC also appears to involve the modification of gut microbiota and DNA demethylation(Reference Chen, Jiang and Zhong39). In this study, we have demonstrated for the first time that the chemopreventive effect of BRB anthocyanins against CRC might also involve the up-regulation of miR-24-1-5p, and we have shown this through the use of a mouse model.
In summary, miR-24-1-5p might act as a tumour suppressor and modulate the growth of CRC cells through down-regulating the expression of β-catenin and its downstream target genes, and this could be part of the mechanism by which BRB anthocyanins manifest its chemopreventive effect against CRC. miR-24-1-5p could therefore be considered as a potential agent for the prevention or treatment of CRC in future.
Acknowledgements
This work was financially supported by National Science Foundation of China to X. B. (grant nos 81272333 and 81472821); programme of Liaoning Excellent Talents in University to X. B. (LETU no. LR 2014001) and in part from Innovation Team Project (no. LT2015011) from the Education Department of Liaoning Province. The authors thank Dr Alan K. Chang for revising the language of the manuscript.
H. Z. and J. G. designed, performed, analysed experiment and wrote the manuscript. L. M., Q. L., M. G., T. M. and Q. Z. performed and analysed experiments. X. B. conceived the study, designed experiments, reviewed and analysed data, and wrote and edited the manuscript.
The authors declare no competing financial interest.










