INTRODUCTION
The microbial communities from icy habitats such as polar ice sheets (Skidmore and others, Reference Skidmore, Foght and Sharp2000), alpine glaciers (Branda and others, Reference Branda2010), Arctic glaciers (Adams and others, Reference Adams, Crump and Kling2014) and permafrost (Frey and others, Reference Frey2016), constitute one of the Earth's biomes (Anesio and Laybourn-Parry, Reference Anesio and Laybourn-Parry2012). Among these, ice caves are one of the least investigated cold habitats regarding the diversity and ecological role of ice-contained microbiomes (Purcarea, Reference Purcarea, Perșoiu and Lauritzen2018). Microorganisms from perennial ice accumulated in caves became of interest during the last two decades with the isolation of novel cold-adapted bacterial species (Margesin and others, Reference Margesin, Gander, Zacke, Gounot and Schinner2003) and the investigation of microbial diversity from ice caves, focused on bacterial communities (Hillebrand-Voiculescu and others, Reference Hillebrand-Voiculescu2013, Reference Hillebrand-Voiculescu2014; Iţcuş and others, Reference Iţcuş, Pascu, Brad, Perşoiu and Purcarea2016, Reference Iţcuş2018), while only addressing eukaryotic microorganisms based on the 18S rRNA gene (Hillebrand-Voiculescu and others, Reference Hillebrand-Voiculescu2014; Brad and others, Reference Brad2018) and internal-transcribed spacer (ITS) (Ogórek, Reference Ogórek2018) sequences. Recent studies revealed the presence of fungal communities in different frozen habitats such as permafrost (Ozerskaya and others, Reference Ozerskaya, Kochkina, Ivanushkina, Gilichinsky and Margesin2009), cryoconites (Edwards and others, Reference Edwards2012), subglacial ice (Sonjak and others, Reference Sonjak, Frisvad and Gunde-Cimerman2006) and Antarctic and high-Arctic glaciers (Cantrell and others, Reference Cantrell, Dianese, Fell, Gunde-Cimerman and Zalar2011). Moreover, the occurrence of particular fungal communities in ice contained caves was mentioned in lava tubes from Mount Erebus, Antarctica (Connell and Staudigel, Reference Connell and Staudigel2013).
Ice caves are secluded and highly preserved cold habitats representing a new source of cold-adapted microorganisms. The morphology and specific climatic conditions of these caves allowed the accumulation of perennial underground ice deposits constituting paleoclimate archives (Perșoiu and Lauritzen, Reference Perșoiu and Lauritzen2018). Therefore, the ice-entrapped microorganisms could be used as proxies for climate variation studies, while the response of the microbiome composition and abundance to changes in temperature and precipitation regimes during ice formation is expected to reflect the impact of climatic variations on ice depositional processes (Liu and others, Reference Liu2016).
Scărișoara Ice Cave (Romania), hosting the largest and oldest (>10 000 years) underground glacier in the world (Perșoiu and others, Reference Perșoiu and Pazdur2017), constitutes a model for paleoclimate studies in ice caves (Perşoiu and others, Reference Perşoiu2017) as well as of the diversity, resilience and ecological role of cave ice microbiome (Purcarea, Reference Purcarea, Perșoiu and Lauritzen2018), in order to understand the impact of climate variations on the microbial community structure and activity in this type of glacial habitat. Investigation of the microbiome from the ice block of Scărișoara Ice Cave revealed the presence of complex uncultured (Hillebrand-Voiculescu and others, Reference Hillebrand-Voiculescu2014; Iţcuş and others, Reference Iţcuş2018) and cultured (Iţcuş and others, Reference Iţcuş, Pascu, Brad, Perşoiu and Purcarea2016) bacterial communities. Cultured fungi were also identified in various cave ice strata using a polymerase chain reaction (PCR)-denaturing gradient gel electrophoresis (DGGE) approach (Brad and others, Reference Brad2018). Although fungi appeared to retain viability for at least a thousand years in ice deposits (Abyzov, Reference Abyzov and Friedman1993; Ma and others, Reference Ma, Catranis, Starmer and Rogers1999), culture-dependent techniques failed to detect extensive fungal community taxa, while the implementation of molecular techniques allowed the identification of these microbial communities at genus and species levels (Ghosh, Reference Ghosh2017). Although the ice geochemistry and climate variations during ice deposition are known to affect the microbiome distribution in various glacial habitats, little is known on the effect of these parameters on environmental fungal communities from perennial cave ice.
This work addresses the lack of knowledge on the temporal distribution of environmental fungal communities in perennial cave ice deposits by investigating the composition of ice-entrenched fungal communities in up to 1500 years old ice layers of Scarisoara cave ice block based on Illumina MiSeq sequencing of the ITS2 region of ribosomal RNA genes, in relation to their geochemical configuration and climate-associated records. To the best of our knowledge, this is the first report on the distribution of environmental fungal diversity in perennial cave ice using high-throughput sequencing.
MATERIALS AND METHODS
Site description and sample collection
Scărișoara Ice Cave (Fig. 1) is located in NW Romania (46°29′23.64″ N, 22°48′37.68″ E) (Holmlund and others, Reference Holmlund2005). This limestone cave, 105 m deep and 700 m long, is a single entrance descending cavity located at an altitude of 1165 m a.s.l. (Fig. 1a). The entrance (60 m in diameter, 48 m deep) gives access to a large chamber (Great Hall) (Fig. 1b). The perennial ice block forms the floor of the Great Hall area. A reduced section of the ice-block surface (~10 m2) in the vicinity of the entrance shaft is exposed to sunlight, containing an abundant phototrophic community (Hillebrand-Voiculescu and others, Reference Hillebrand-Voiculescu2014). Melting and sublimation due to geothermal heat and cold air circulation led to a gradual retreat of the sides of the ice block on the northern part of the Great Hall, resulting in the development of a vertical cliff, where ice layers deposited during the last millennium are visible (Perşoiu and Pazdur, Reference Perșoiu and Pazdur2011). The ice block was mainly formed by the annual freezing in late autumn of ponded drip, rain/snowmelt and seepage water, overlapping the ice from the previous year (Racoviță, Reference Racovita1927; Perşoiu and others, Reference Perşoiu, Onac and Perşoiu2011a). Since drip water is carrying large amounts of organic matter (e.g. soil, pollen, invertebrates), the freezing process resulted in the genesis of a varve-like deposit, comprising both clear ice (1–20 cm thick) and sediment-rich layer (Fig. 1c), including organic material (Racoviță and Onac, Reference Racoviță and Onac2000; Feurdean and others, Reference Feurdean2013) and cryogenic precipitated calcite (Žák and others, Reference Žák, Onac and Perşoiu2008). The morphology of the cave and the presence of ice determine a peculiar climate (Racoviță, Reference Racoviță1994; Perşoiu and others, Reference Perşoiu, Onac and Perşoiu2011a), with cold air inflow in winter and thermal stratification in summer, with a layer of warm air floating on the denser, colder air inside the cave. The distal parts of the cave, unglaciated, have positive temperature year-round, while those in the vicinity of the entrance (where the ice block is also present) have temperature variations that follow those outside the cave in winter and stay stable at 0 °C during summer.
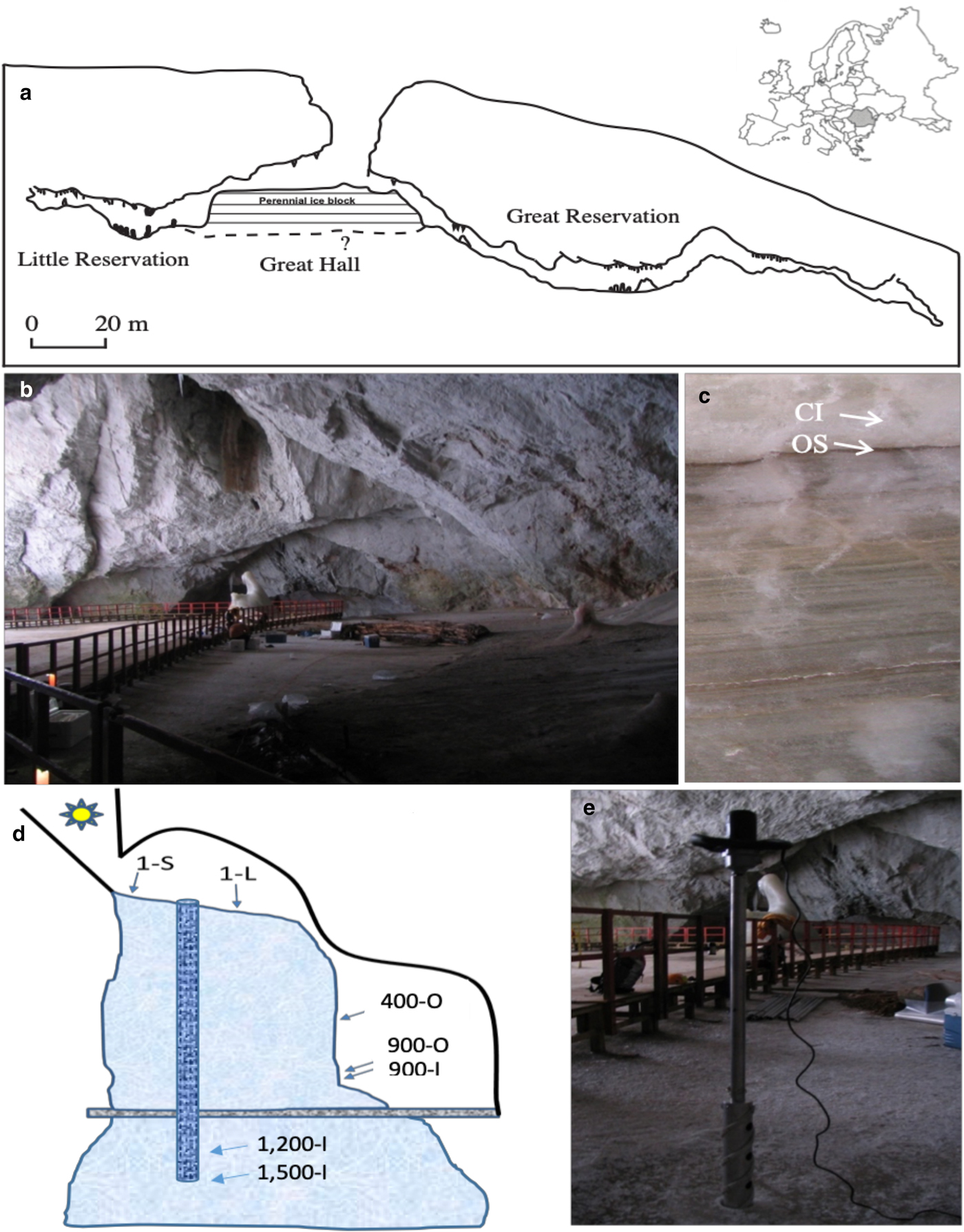
Fig. 1. Scărișoara Ice Cave sampling. (a) Cave map indicating the location of the perennial ice block of undetermined (?) depth (modified from Perșoiu and others, Reference Perşoiu, Onac, Wynn, Bojar and Holmgren2011b). (b) Great Hall ice floor constituting the top of the underground ice block (1-S and 1-L ice samples) (photos: C. Purcarea). (c) Little Reservation Ice wall showing horizontal stratification with clear ice (CI; sample 900-I) and organic sediment (OS; samples 400-O, 900-O). (d) Schematic representation depicting Scărișoara Ice Cave and ice-block morphology with sampling sites annotation. (e) Vertical drilling of the ice block in the Great Hall area.
Various geological (Racoviță and Onac, Reference Racoviță and Onac2000) and paleoclimatic (Perșoiu and Pazdur, Reference Perșoiu and Pazdur2011; Perșoiu and others, Reference Perşoiu2017) studies showed that the perennial ice block inside the cave has accumulated for millennia, containing a high (decadal) resolution record of climate and environment changes during the last 1500 years, including the well-known cold and dry Little Ice Age (LIA, AD 1250–1860) and Dark Ages Cold Period (DACP, AD 400–800), and the warm and wet Medieval Warm Period (MWP) (AD 800–1250) (Perșoiu and Pazdur, Reference Perșoiu and Pazdur2011; Perșoiu and others, Reference Perşoiu2017).
Samples collection
Ice samples were collected from seven different sites within the ice block (Fig. 1d) corresponding to various deposition periods encompassing recent, LIA and MWP intervals, and from strata of high and low organic sediment content, in order to correlate the fungal community distribution in this glacial habitat with paleoclimate records and ice geochemistry. Recently-formed (1 year old) ice resulted from the freezing of seepage and rain/snowmelt water accumulated was collected from the surface of the glacier in the Great Hall. Sample 1-S was collected from a sun-exposed cave site near the entrance, while sample 1-L was collected from a similarly-formed ice layer but from an indirect sunlight exposed area in the centre of the Great Hall (Fig. 1a). These samples were collected from the top of the ice block by vertical drilling (Fig. 1e). The 400 (sample 400-O), and 900 (samples 900-O and 900-I) years-old ice samples were collected from the ice wall of Little Reservation (Fig. 1c) by horizontal drilling, after removing ~20 cm of the surface ice, while 1200 (sample 1200-I) and 1500 (sample 1500-I) years old ice samples were collected from the ice core obtained by vertical drilling in the central area of Great Hall (Fig. 1e). The age of samples 400-O, 900-O and 900-I were determined by 14C dating of macrofossils (leaves and tree branches) collected from the same layers as the samples collected for microbiological analyses (Perșoiu and Pazdur, Reference Perșoiu and Pazdur2011), while the ages for samples 1200-I and 1500-I were determined by Bayesian interpolation (Blaauw and Christen, Reference Blaauw and Christen2011) between 14C ages of organic matter trapped in ice (Perșoiu and others, Reference Perşoiu2017) (Table S1). Samples collected from organic-rich ice layers were labelled with ‘O’ (400-O, 900-O), and ‘I’ (900-I, 1200-I, 1500-I) for those collected from a layer of clear ice, with no visible organic content (Fig. 1c). Sampling was carried out in triplicate from each location, under aseptic conditions, as previously described (Hillebrand-Voiculescu and others, Reference Hillebrand-Voiculescu2014). The ice surface was flamed for 5–10 s prior to the drilling procedure, and both the outer and inner surfaces of the coring auger were decontaminated with 96% ethanol and flaming after each drilling step. Ice samples were collected in sterilised flasks, in the presence of an open-flame laboratory torch.
Geochemical analyses
Ice samples were melted at 4 °C and filtered on 0.22 µm MF-membranes (Millipore). The pH and electric conductivity were measured using a S40 SevenMulti™ pH/mV meter (Mettler Toledo, USA). Dissolved total carbon (DTC), dissolved inorganic carbon (DIC) and dissolved organic carbon (DOC) contents were determined as previously described (Iţcuş and others, Reference Iţcuş, Pascu, Brad, Perşoiu and Purcarea2016) using a Multi N/C 3100 elemental combustion analyser (Analytic Jena, Germany). Element determinations in melted and filtered water (Hydrogeochemistry Laboratory, Emil Racoviţă Institute of Speleology, Bucharest) were performed by inductively-coupled plasma mass-spectrometry using a NexION 300S system (PerkinElmer, Shelton, CT, USA), in accordance with US-EPA (2014) 6020B standards. Ca concentrations were determined in dynamic reaction cell mode by using NH3 as a reactive gas. Na, K, Mg, Sr, Cu and Zn concentrations were measured in kinetic energy discrimination mode with He as an inert gas, while Cl concentrations were determined using standard mode (US-EPA, 2014). Calibration solutions were purchased from High-Purity Standards™ (Charleston, SC, USA). For the laboratory control samples, NIST Standard Reference Material®1640a was used for measuring Na, K, Mg, Ca, Sr, Cu and Zn contents, and Simulated Seawater Standard (HighPS) for chloride measurements. The uncertainty of the analytical measurements was estimated according to ISO 11352:2012 standard.
DNA extraction
Ice samples (200–300 mL) were thawed at 4 °C and filtered on 0.22 µm MF-membranes (Millipore) using a vacuum-driven stainless-steel filtering system (Millipore) and Chemical Duty Pump, 220 V/50 Hz (Merck Millipore), under sterile conditions. Filters containing biomass were cut into small pieces, and the total microbial DNA was extracted using a Blood and Tissue DNA extraction kit (Qiagen) by a modified protocol including two initial cell lysis steps. After incubation for 1 h at 37 °C in the presence of 200 µL TE containing 20 units mutanolysin (Fermentas) per g of filtered cell biomass, to disrupt the microbial cell wall, the cell suspension was incubated for 30 min at 56 °C in the presence of 12 units proteinase K per g of biomass and ZR bashing beads (Zymo Research), using a Homogenisation system SpeedMill PLUS homogeniser (Analytic Jena), and further cell lysis performed by two cycles of 3 min on interspersed by two cycles of 3 min off in order to prevent sample overheating. DNA quantitation assays were assessed using a fluorescence-based Qubit® 2.0 Fluorometer (ThermoFisher Scientific).
Library preparation and Illumina MiSeq sequencing
PCR were performed to amplify the ITS2 region of the eukaryotic ribosomal operon for the gDNA samples. Each reaction was performed using three different dilutions of the samples (1/50, 1/100, 1/2000) using a Taq 5U-ul Qiagen HotStarTaq with specific primers (Frey and others, Reference Frey2016) ITS3-CS1 (ACACTGACGACATGGTTCTACACAHCGATGAAGAACGYRG) and ITS4-CS2 (TACGGTAGCAGAGACTTGGTCTTCCTSCGCTTATTGATATGC) (CS1 and CS2 tags are underlined). Each DNA fragment was amplified using a PCR thermal cycler (DNA Engine; Bio-Rad, Hercules, CA, USA) with the following thermal cycling conditions: the first cycle consisted of 15 min at 96 °C, followed by 33 cycles of 30 s at 96 °C, 30 s at 52 °C for annealing, 1 min at 72 °C, and a final incubation for 10 min at 72 °C. Progressively, after the verification of the PCR products on 2% agarose gel, the barcoding step was performed adding a barcode to each sample and the sequence of Illumina adapters. For barcoding, 1/50 dilution of the PCR reactions were used with the following thermal cycling conditions: the first cycle consisted of 10 min at 95 °C, followed by 15 cycles of 15 s at 95 °C, 30 s at 60 °C for annealing, 1 min at 72 °C, and a final incubation for 3 min at 72 °C. The resulting barcoded amplicons were sequenced using the Illumina MiSeq PE250 bp platform (Génome Québec Innovation Centre, Canada) with the MiSeq Reagent Kit v2 500 cycles from Illumina.
Sequence quality control, operational taxonomic unit (OTU) clustering and taxonomic assignments
Quality control clustering into OTUs and subsequent taxonomic identification were conducted as previously described (Frey and others, Reference Frey2016). A customised pipeline based on UPARSE (Edgar, Reference Edgar2013; Edgar and Flyvbjerg, Reference Edgar and Flyvbjerg2015) implemented in USEARCH v. 9.2 (Edgar, Reference Edgar2010) was used by applying the USEARCH fastq mergepairs algorithm (Edgar and Flyvbjerg, Reference Edgar and Flyvbjerg2015) to merge paired-end reads filtering for a minimum length of 300 bp. The Bayes Hammer algorithm (Nikolenko and others, Reference Nikolenko, Korobeynikov and Alekseyev2013) was used to correct substitution errors from phasing events during Illumina sequencing, and Cutadapt software (Martin, Reference Martin2011) to remove PCR primers, allowing for a maximum of one mismatch in the forward and reverse primer, respectively. Subsequently, the USEARCH fastq_filter function was used for quality-filtering discarding reads with an expected error of one or greater. To remove singletons, the set of dereplicated sequences was clustered into OTUs at 97% identity using the cluster_otu function implemented in USEARCH (Edgar, Reference Edgar2013) that included an ‘on the fly’ chimera removal algorithm. ITSx software (Bengtsson-Palme and others, Reference Bengtsson-Palme2013, Reference Bengtsson-Palme2015) was used to assess the presence of ribosomal signatures. Sequences were subsequently mapped on the OTU centroid sequences. Taxonomic classification was conducted using the naïve Bayesian classifier classify.seqs function (Wang and others, Reference Wang, Garrity, Tiedje and Cole2007) implemented in Mothur (Schloss and others, Reference Schloss2009) with a minimum bootstrap support of 0.6. Sequences were queried against the NCBI database (Sayers and others, Reference Sayers2009), and the OTUs identified as Fungi were further classified using the UNITE database (Nilsson and others, 2015). Nonfungal sequences were removed for downstream analyses. Close relatives of the assigned OTUs were identified using the BLAST (basic local alignment search tool) algorithm and NCBI database (Altschul and others, Reference Altschul, Gish, Miller, Myers and Lipman1990).
The ITS2 raw sequences were deposited in the NCBI Sequence Read Archive under the BioProject accession number SRP158737.
Statistical analyses
Observed richness and Shannon diversity were calculated based on nonrarefied OTU abundance matrices. Differences in α-diversity (local diversity, Whittaker, Reference Whittaker1960) were examined conducting a one-way analysis of variance (ANOVA) in R v.2.15 (R development Core Team, 2012). Pairwise differences among sites were assessed using Tukey's Honestly Significant Difference (Tukey HSD) post-hoc test with the TukeyHSD function (R development Core Team, 2012). Rarefaction curves were calculated using the function rarecurve implemented in the R package vegan. In order to analyse differences in fungal community structures, we calculated Bray–Curtis based on square-root-transformed relative OTU abundances. Statistical evaluation of β-diversity patterns was performed by conducting a permutational ANOVA (PERMANOVA, number of permutations = 9999) with the function adonis and an analysis of similarity (ANOSIM, number of permutations = 9999) implemented in the vegan R package (Oksanen and others, Reference Oksanen2007). Monte Carlo pairwise comparisons of fungal community structures were conducted using Primer6+. Principal coordinate analysis (PCoA) ordinations were calculated using the ordinate function implemented in the R package phyloseq (McMurdie and Holmes, Reference McMurdie and Holmes2013). Geochemical variables were correlated with PCoA ordination scores using the envfit function implemented in the R package vegan (Oksanen and others, Reference Oksanen2007). PCoA and abundance barplots were generated in R with the ggplot2 package (Wickham, Reference Wickham2016) if not specified otherwise.
RESULTS
Cave ice geochemical properties
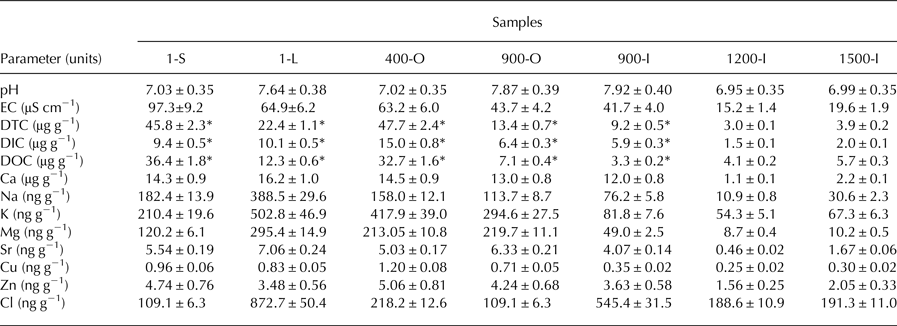
Geochemical parameters of 1-S, 1-L, 400-O, 900-O, 900-I, 1200-I and 1500-I samples comprising pH, conductivity, concentrations of DTC, DIC, DOC and several specific ions (Table 1) were determined in order to analyse the dependence of ice-contained fungal community composition on the (physico)chemical characteristics of ice deposits of different ages from Scarisoara Ice Cave. The pH ranged from 6.95 to 7.92 across the ice block, with neutral values in most strata except for the clear ice 1-L and both 900-O (pH 7.87) and 900-I (pH 7.92) years old samples where the ice deposits were slightly alkaline (7.64–7.92). The electrical conductivity (EC) of melted ice samples showed the highest value (97.3 μS cm−1) for the recently-formed ice 1-S, and an exponential decrease with the age of ice up to 15.2–19.6 μS cm−1 in older ice samples 1200-I and 1500-I (Table 1). DTC was unevenly distributed across the ice block, with high values (45.8–47.7 µg g−1) for the 1-S and 400-O samples, and twofold lower (22.4 µg g−1) in recently formed clear ice 1-L. Meanwhile, older ice strata showed a fourfold (900-O and 900-I) and tenfold (1200-I and 1500-I) more reduced DTC content. The DOC content varied in the 3.3–36.4 µg g−1 range, with higher values for 1-S (36.4 µg g−1) and 400-O (32.7 µg g−1), and up to eightfold decreased contents (3.3– 5.7 µg g−1) in older clear ice strata (Table 1). Among elements, Ca was the dominating ion across the ice block, as expected for a limestone cave deposits (Table 1), with comparable concentrations (14–16.2 µg g−1) in the ice layers formed during the last 900 years, and severely decreased values in older ice samples 1200-I (1.1 µg g−1) and 1500-I (2.2 µg g−1). Na, K, Mg and Cl concentrations were five to tenfold lower than that of Ca, showing a decrease with the age of ice. Relatively high concentrations of Cu, Zn and Sr (0.25–7.06 ng g−1) were also present in all cave ice samples, with two/fivefold diminution in 1200-I and 1500-I strata, respectively. Chloride ion concentration of ~100 ng g−1 was observed in 1-S and 900-O samples, whereas a twofold higher content was found in 400-O, 1200-I and 1500-I samples. Moreover, the clear ice samples 1-L and 900-I had an eightfold and fivefold higher Cl concentration that the sun exposed 1-year old ice (1-S), respectively.
Table 1. Geochemical properties of Scarisoara ice samples. The pH and EC, DTC, DIC and DOC concentrations were measured as indicated in Materials and methods section; *previously determined for 1-S, 1-L, 400-O, 900-O and 900-I samples (Iţcuş and others, Reference Iţcuş, Pascu, Brad, Perşoiu and Purcarea2016)
Fungal abundance and diversity across Scarisoara ice block
Illumina MiSeq sequencing of ITS2 region applied to the triplicate ice samples from Scarisoara ice block up to 1500 years old ice layers led to a total of 1 751 957 sequences ranging in the 62 555–105 357 reads per sample interval, with an average of 83 426.52 reads/sample (33.5 quality) (Table S2). Among the total ITS2 sequences, 64% were assigned to fungal taxa (Fig. 2), while 13% belonged to green algae, 12% to plants (Tracheophyta) and 9% to Alveolates, besides a low (1%) presence of Metazoa and Synurophyceae.
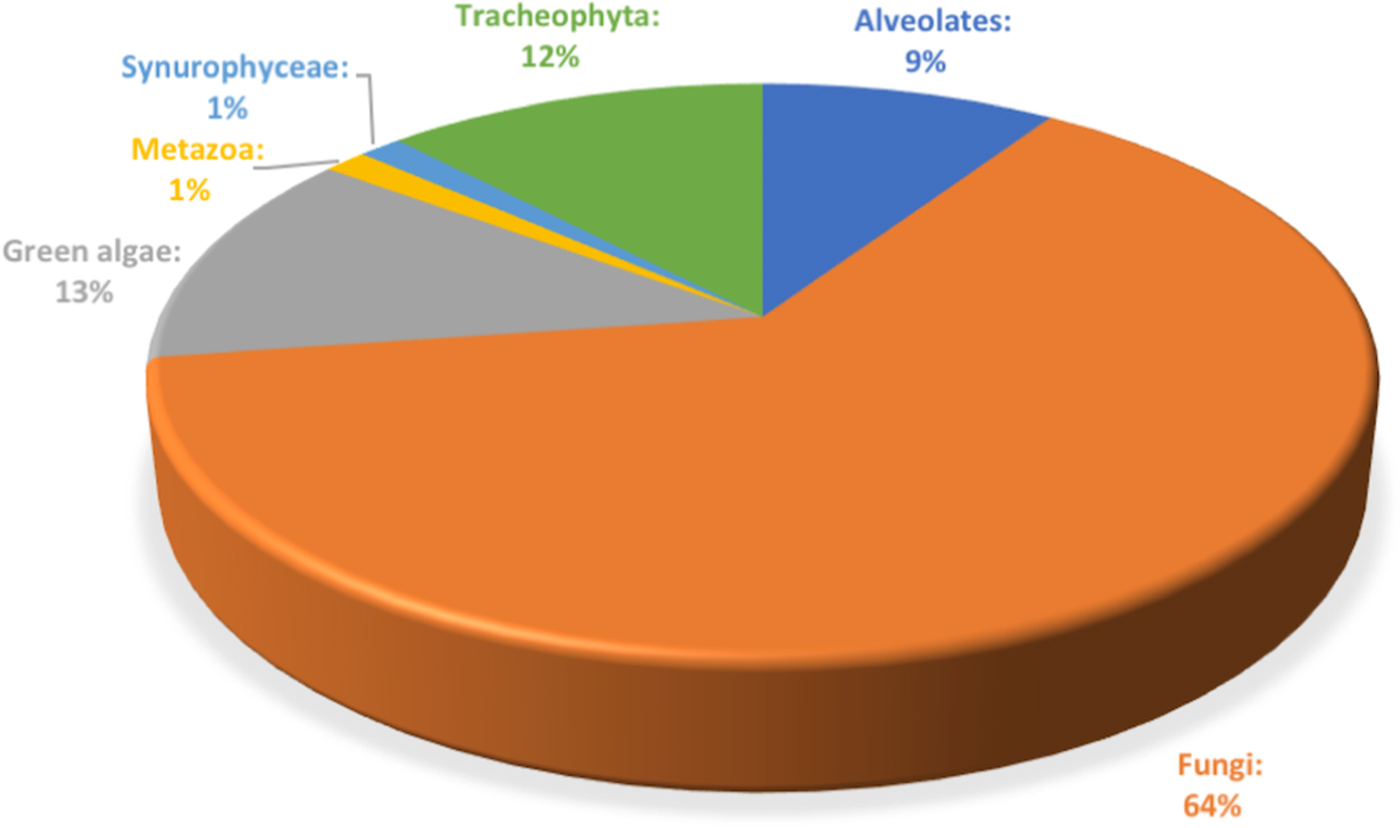
Fig. 2. Relative content of microbial eukaryote ITS2 amplicons identified across the 1500 years Scarisoara ice block.
After eliminating the nonfungal ITS2 sequences, the resulting 182 fungal OTUs were assigned to four phyla, 16 classes, 37 orders, 63 families and 80 genera. Ascomycota was overall the most abundant (30–90%) phylum across the ice block, followed by Basidiomycota (5–25%), Chytridiomycota (20%) and Mucoromycota (1%) (previously known as Zygomycota; Spatafora and others, Reference Spatafora2016), with 20% unclassified fungal taxa. The rarefaction curve (Fig. S1) indicated a clearly visible saturation for the fungal communities in older ice strata (400-O, 900-I, 1200-I and 1500-I), associated with a low diversity in these samples, in particular for 1200-I and 1500-I. Recently formed ice 1-S and 1-L had a higher number of species (up to 70%), with large variation between the replicates. No saturation was reached in cases of 900-O and 1200-I, even after 60 000 reads (sample size), indicating a partial identification of fungal community composition for these cave ice strata.
The distribution of fungal OTUs in different aged ice strata (Fig. 3) revealed a heterogeneous phylotype occurrence across the cave ice block. Two of the OTUs representing 1.1% of the fungal community were common to all samples, and 28 OTUs were shared between ice layers of different ages, while 153 OTUs were specifically found in distinct cave ice deposits. The highest number (119) of unique fungal phylotypes, representing 65.4% of the fungal community, was found in recent ice samples 1-S and 1-L, while the older ice contained 2.7% (400-O), 11% (900-O and 900-I) and 3.8% (1500-I) less ITS2 amplicons, and only two specific OTU in the 1200-I ice sample (1.1%) of total identified fungal phylotypes.
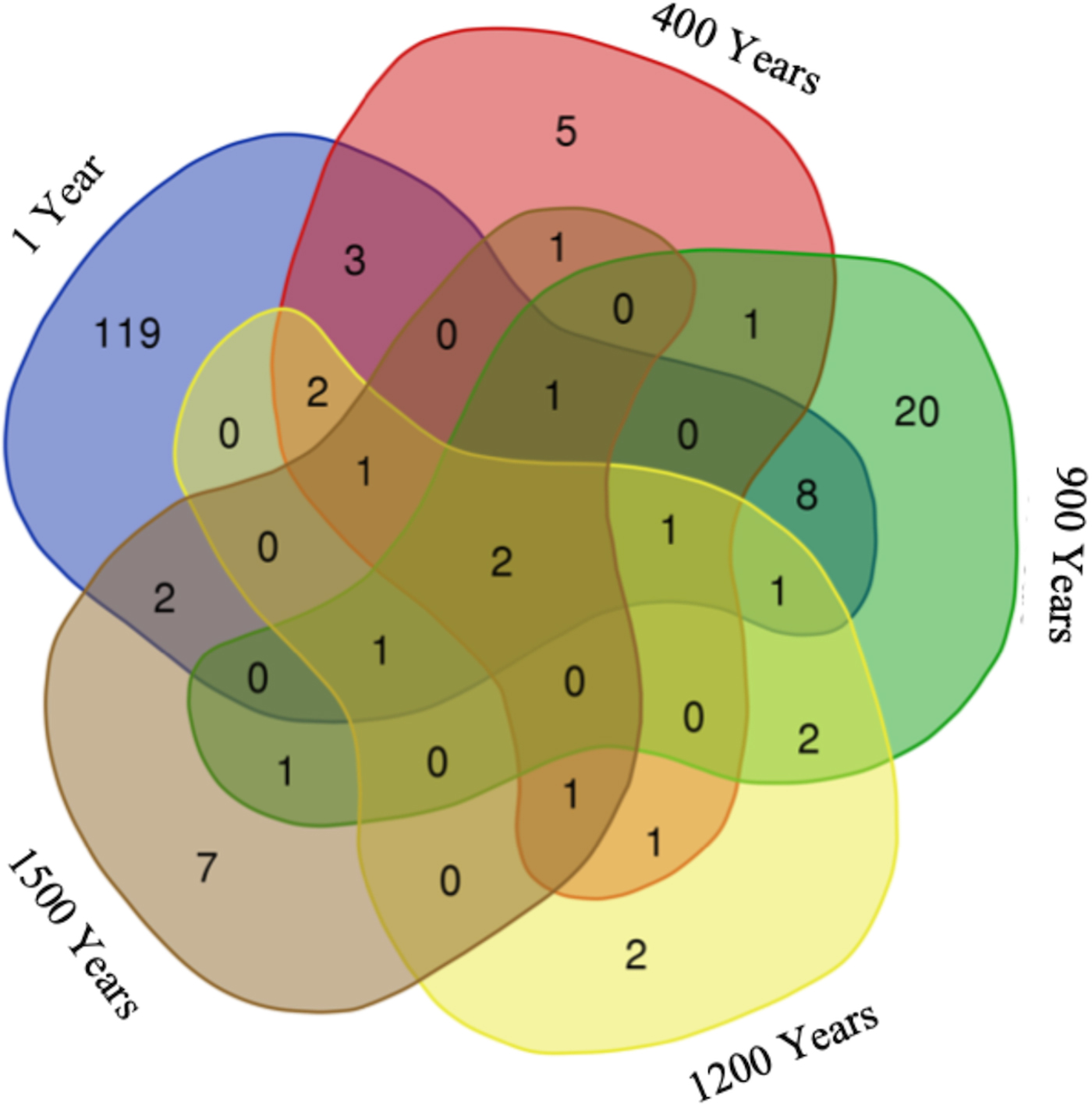
Fig. 3. Distribution of fungal OTUs across Scarisoara ice block. The VENN diagram indicates the number of unique and shared ITS2 OTUs in 1, 400, 900, 1200 and 1500 years old cave ice layers.
The two fungal OTUs shared among all ice-block layers (Fig. 3) were assigned to Cryptococcus victoriae and ANTELF16 unclassified fungal species. Recently formed ice deposits (1 year) contained 119 unique OTUs (mostly belonging to the Ascomycota phylum) in which the dominant species comprised Leucosporidiales, Capronia and Helotiales genera, with the presence of two uncultured Cystofilobasidiales and Rhinocladiella sp. Among specific fungal OTUs of 400 years old ice we identified five Mortierella OTUs. In 900 years old ice, 20 unique OTUs belonging to the Basidiomycota phylum were assigned to Rhodotorula, Schizopora and Dioszegia genera. The two OTUs found only in 1200 years old ice corresponded to Chaetothyriales and Agaricomycetes species, while the seven unique OTUs from 1500 years old ice were assigned to Dothideomycetes, Eurotium, Phaeosphaeria, Itersonilia and Fomes species. The shared fungal community between 1 and 900 years old ice strata contained eight members of the Ascomycota, Basidiomycota and Chytridiomycota phyla. Only three OTUs, all belonging to the Ascomycota phylum with one assignment at the genus level (Cadophora sp.), were shared between 1 year and 400 years old ice layers, while a single OTU, assigned to Lyophyllum connatum sp. belonging to Basidiomycota, was commonly present in 400 and 900, and 1200 years old ice samples.
α-diversity
The diversity of the identified fungal community from Scărișoara ice block was relatively low (Fig. 4). Observed richness ranged between 3 and 69 (Fig. 4a) while Shannon diversity ranged between 0.1 and 3.2 (Fig. 4b). Both observed richness and Shannon diversity were significantly (P < 0.05) higher in samples 1-L and 1-S as compared to older ice layers, while no significant difference was found among the older ice samples 400-O, 900-O, 900-I, 1200-I and 1500-I (Table 2).

Fig. 4. α-diversity indices for fungal OTUs identified in Scărișoara ice block. (a) Observed OTUs and (b) Shannon index variations were measured from triplicate assigned OTUs of 1-S, 1-L, 400-O, 900-O, 900-I, 1200-I and 1500-I ice samples, using the nonrarefied sequences (McMurdie and Holmes, Reference McMurdie and Holmes2014). Letters (a, b) indicate the significance difference level at P < 0.05. Horizontal lines represent the median, while the boxes represent the interquartile range of the first and third quartiles. The vertical lines (whiskers) represent the maximal and minimal values.
Table 2. Statistical differences in α-diversity and β-diversity of fungi in the different analysed ice layers
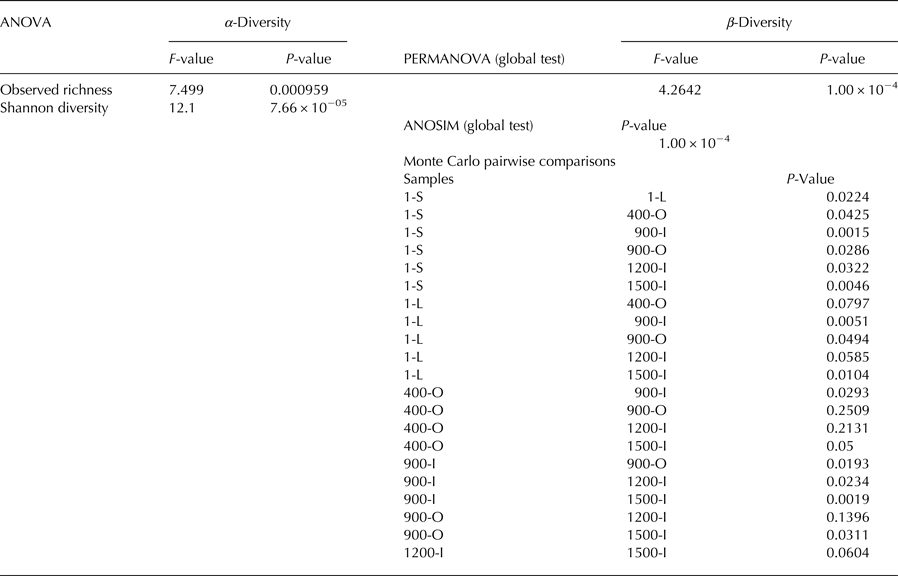
α-diversity: a one-way ANOVA was conducted to assess differences by site in observed richness and Shannon diversity followed by Tukey's Honestly Significant Difference (Tukey HSD) post-hoc test. β-diversity: PERMANOVA and ANOSIM were performed to investigate differences in fungal community structures based on Bray–Curtis distances of square-root-transformed relative abundances. Monte Carlo test was used to assess pairwise differences in community structures between sites.
Fungal community structure
Illumina ITS2 sequencing revealed an unevenly distributed fungal community throughout the ice block (Fig. 5). The relative abundance of fungal phyla (Fig. 5a) indicated that Ascomycota was the major phylum in the cave ice layers formed during the last 1500 years, dominating the older ice samples 900-O, 1200-I and 1500-I (75–80%), and highly represented in 400-O (48%), 1-L (60%) and 1-S (30%) ice samples. Meanwhile, this phylum was scarcely distributed (7%) in the 900-I sample. OTUs belonging to Basidiomycota were present in all strata formed during the last millennium, ranging from 5 to 10% in all ice layers, except for the 900-I clear ice sample that appeared highly dominated (90%) by this taxa and 1500-I sample where it was absent (Fig. 5a). Chytridiomycota phylum was found only in recent ice samples, with higher relative content (25%) in the direct sunlight exposed ice 1-S, while barely (<1%) present in the 1-L sample. OTUs within the Mucoromycota phylum were identified only in the 400-O sample (<1%). Most of unclassified fungal OTUs (25%) were present in both recent ice 1-S and 1-L, in 400-O (30%), and, to a small extent (4%), in the 900-O ice sample.
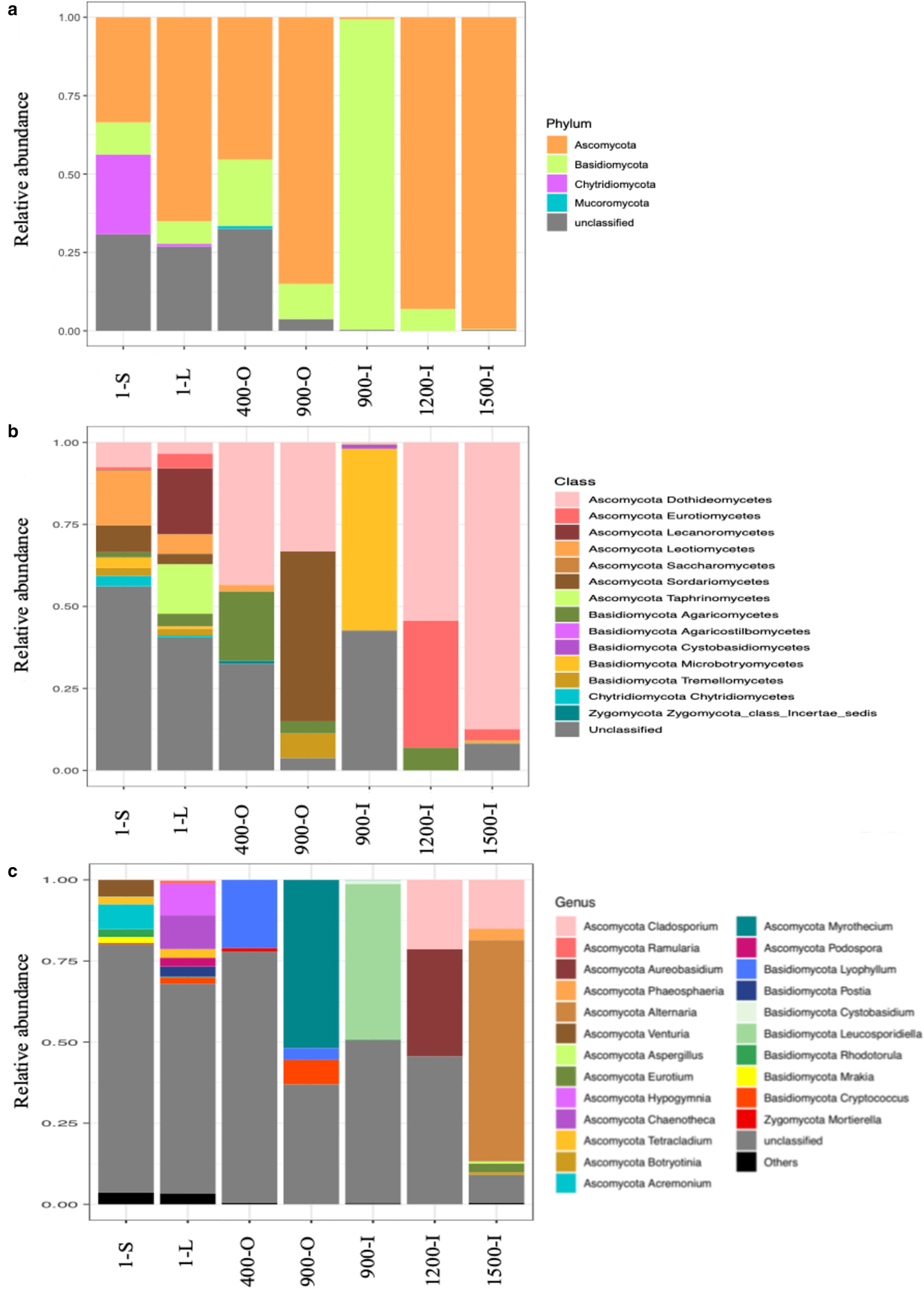
Fig. 5. Relative abundance of the dominant fungal phyla (a), classes (b) and genera (c) in 1-S, 1-L, 400-O, 900-O, 900-I, 1200-I and 1500-I ice samples from Scărișoara Ice Cave based on ITS2 Illumina sequencing.
At the class level (Fig. 5b), Dothideomycetes were found at high relative content (90%) in older ice 1500-I, and with only low contents (4–10%) in recent ice 1-S and 1-L samples. The occurrence of this fungal class appeared to decrease in diversity and increase in relative abundance in older ice strata. Overall, a higher relative abundance of classes belonging to the Ascomycota phylum was found in each sample except for 900-I where Microbotryomycetes (Basidiomycota) was the dominant fungal class (60%). Sordariomycetes (Ascomycota) OTUs were identified in recently formed ice 1-S and 1-L (4–10%), while barely present (0.5%) in the 400-O sample and absent in older ice samples (900-I, 1200-I and 1500-I). However, a high representation (50%) of this class was found in the 900-O sample characterised by higher organic matter content. Eurotiomycetes were mainly present in the 1200-I ice (40%), and scarcely distributed in the 1-L sample (5%), while absent in the remaining ice layers. The unclassified fungal taxa occurred to a large extent (30–55%) in recent ice 1-S and 1-L, and in 400-O samples, respectively, while only scarcely represented in the 900-O and 1200-I samples (0,5–2%). Interestingly, while the unclassified fungi were barely identified in 900-O and completely absent in the 1200-I ice layer, the 900-I sample was characterised by a high (45%) content of unidentified fungi.
At the genus level (Fig. 5c), 1-S and 1-L and 1500-I ice samples exhibited a relatively high diversity, while 400-O, 900-O, 900-I and 1200-I ice layers had a lower fungal variability. Lyophyllum sp. was the most abundant genus in the 400-O sample. Myrothecium sp., belonging to Ascomycota, had the highest relative abundance in organic-rich 900-O ice (52%), while Leucosporidiella sp. (Basidiomycota) constituted the most represented species in clear ice 900-I of the same age. In the 1200-I sample, the most abundant species belonged to Cladosporium and Aureobasidium genera (Ascomycota). An unexpected higher diversity was found in the oldest ice sample 2000-I, counting five genera, with Cladosporium sp. and Alternaria sp. representing 15 and 75%, respectively, of fungal species. Chytridiomycota phylotypes were identified in recently formed ice strata 1-S and 1-L belonging to Rhizophydium sp. and Spizellomycetales sp.
A high relative content of unclassified fungal taxa was found across the cave ice block (25%), with a higher representation in recently formed ice and decreased with the age of ice, suggesting the presence of a significantly higher fungal diversity in the cave glacier, in particular in younger ice samples. However, this high percentage of unclassified taxa could also be due to DNA degradation in these ice strata, as well as the uncertainty in fungal sequence identification.
β-diversity and geochemical impact on fungal community structure
A PCoA analysis of fungal OTUs across the cave ice block in relation to the geochemical parameters (Fig. 6) showed differences in β-diversity explaining 32.3% of variation in the community structures with 16.9% (PCO1 axis) and 15.4% (PCO2 axis), respectively. Fungal community composition mostly followed the age gradient of the ice samples, with 1-year old ice forming a very distinct cluster, while community structures from older strata were separated to a smaller extent. Sample 900-I, however, did not follow this pattern but clustered apart from all other samples.
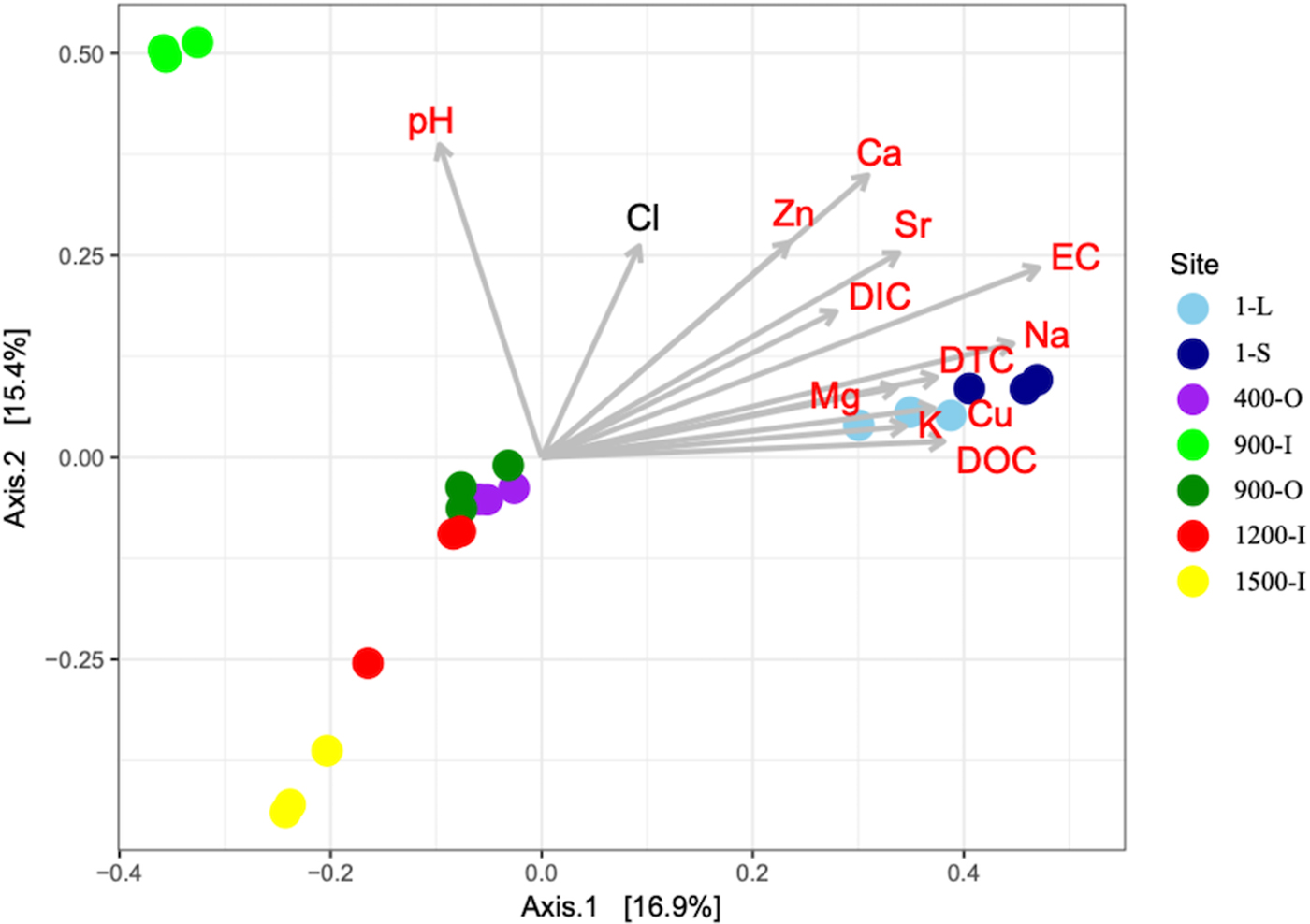
Fig. 6. PCoA of fungal community structure from 1-S, 1-L, 400-O, 900-O, 900-I, 1200-I and 1500-I cave ice samples based on Bray–Curtis dissimilarities in the seven icy habitats and their correlations with geochemical variables. The variability of the PCoA axes is given in parenthesis.
A PERMANOVA analysis revealed significant differences between the sites (P = 1 × 10−04) which was supported by ANOSIM analysis (F = 4.2642). For ice deposits of the same age, Monte Carlo pairwise comparisons revealed significant differences of fungal community structures between 900-O and 900-I samples, whereas a smaller difference was found between the 1-L and 1-S samples. Interestingly the two 900-O and 400-O samples clustered closely together (Table 2), indicating an effect of geochemical composition on the fungal community composition of organic-rich ice deposits from Scarisoara Cave.
All geochemical variables except for Cl were significantly correlated with the ordination scores of the PCoA of fungal community structure (Fig. 6). EC, Ca, Na and Sr were the variables displaying the strongest correlations (R 2 = 0.7686, 0.6039, 0.6086 and 0.4959 respectively) and thus best explaining variance in the fungal community structures (Table S3). Most variables were negatively correlated with ice age whereas pH was only correlated with the differences in fungal community structures in 900-I. Moreover, the carbon content had a major effect on the fungal diversity in the cave ice block, indicating a clear clustering of the recent and older ice strata.
DISCUSSION
Distinctive ice-entrapped fungal community in 1500-years old cave ice block
As a new source of biodiversity, Scărișoara Ice Cave microbiome has been investigated during the last decade. These studies, performed by both traditional culture-dependent and cultured-DGGE methods applied to cultured microorganisms (Iţcuş and others, Reference Iţcuş, Pascu, Brad, Perşoiu and Purcarea2016; Brad and others, Reference Brad2018), revealed the presence of fungi in the perennial ice block harboured in this cave. To overcome the limitations of these applied techniques, recent studies have demonstrated the potential of ITS regions sequencing for quantifying and characterising the fungal diversity in special biological samples (Xia and others, Reference Xia2016). In order to assess a deeper fungal diversity across the ice block from Scărișoara Ice Cave, we used Illumina MiSeq sequencing of ITS2 amplicons. This approach allowed a direct comparison with fungal community structure from other cold environments such as alpine permafrost (Frey and others, Reference Frey2016) and alpine glaciers (Branda and others, Reference Branda2010; Rime and others, Reference Rime, Hartmann and Frey2016).
Our data revealed a nonhomogeneous distribution of fungal taxa throughout Scărișoara Ice Cave layers depending on the age of ice and geochemistry. The identified 182 fungal OTUs were assigned to four phyla, 16 classes, 37 orders, 63 families and 80 genera of fungi. Ascomycota, containing parasitic fungi virtually found in all terrestrial and aquatic ecosystems (Schoch and others, Reference Schoch2009) and playing a major role in recycling dead plant material, was the most abundant (30–90%) phylum across the perennial ice accumulated during the last 1500 years, with an increasing trend in older ice strata, followed by Basidiomycota (5–25%), Chytridiomycota (20%) and Mucoromycota (1%), with 20% unclassified fungal taxa. Basidiomycota, common plant pathogens dominating a large variety of cold environments (Buzzini and others, Reference Buzzini, Turk, Perini, Turchetti, Gunde-Cimerman, Buzzini, Lachance and Yurkov2017) due to their specific adaptation mechanisms to low temperatures (Vishniac, Reference Vishniac, Rosa and Gabor2006), occupied a much smaller share of their cave ice fungal community. Chytridiomycota representatives, known as algae and plants parasites associated with liquid water (James and others, Reference James2006), were present in recent cave ice (1-S), in accordance with the high abundance of algae in the sun-exposed supraglacial pond formed during the summer period (Hillebrand-Voiculescu and others, Reference Hillebrand-Voiculescu2014). Interestingly, Scărișoara Ice Cave deposit represents, to the best of our knowledge, the first icy habitat reported to harbour this fungal phyla. The cave ice block contained Mortierella sp. (Mucoromycota) known as opportunistic fungi, commonly found in other icy habitats (Frӧhlich-Nowoisky and others, Reference Frӧhlich-Nowoisky2015). Interestingly, the relative abundance of unclassified fungi appeared to be higher in cave ice strata formed during the last 400 years that could be related to their presence in this ice layer mainly as spores (allegedly parasitic fungi) which are more resistant under frozen conditions, or might correspond to DNA degradation during ice formation. The spore formation could constitute an adaptation mechanism to frozen environments considering the formation of the cave ice deposits by winter freezing of the supraglacial pond accumulated on top of the ice block (Perşoiu and others, Reference Perşoiu, Onac and Perşoiu2011a) entrapping microbes that could survive for millennia (Gould, Reference Gould2006).
A BLAST analysis of the most abundant OTUs from each ice age deposition (Table S4) indicated the dominance of Ascomycota homologues (>97% identity; E-value >5 × 10−153; query cover >98%) in all strata in addition to Chytridiomycota in recently formed ice, with six of the closest match phylotypes showing a lower (92–96% identity; E-value >2 × 10−94; query cover >96%) identity percentage. These fungi are closely related to species majorly retrieved from plants, particularly trees, rotten/dead wood and soil. Basidiomycota taxa, also widespread throughout the cave ice block (Table S4) was highly represented in 900 years old ice by Leucosporidiella yakutica, Sporidiobolales sp. and Myrothecium sp. (100% identity) also found in China permafrost, plants (Germany), and soil (USA), respectively. Among these taxa, the psychrophilic yeast belonging to Leucosporidium sp., Sporobolomyces sp. and Malassezia sp. were also dominating the fungal communities of saline Antarctic brines from Northern Victoria land (Borruso and others, Reference Borruso2018), showing their potential to adapt to (poly)extreme environments. As compared to other icy habitats, a unique fungal community was identified in the perennial ice strata of Scărișoara Ice Cave. Basidiomycota appeared to be the dominant taxa fungal communities from glacial environments including Antarctic lake (Gonçalves and others, Reference Gonçalves, Vaz, Rosa and Rosa2012), Antarctic (Abyzov, Reference Abyzov and Friedman1993), Arctic (Butinar and others, Reference Butinar, Spencer-Martins and Gunde-Cimerman2007; Dong and others, Reference Dong2016) and alpine (Branda and others, Reference Branda2010; Rime and others, Reference Rime, Hartmann and Frey2016) glacier ice isolated from different locations of Antarctica, Europe, USA, China, India and New Zealand. In comparison with Scarisoara cave ice, the fungal community from the sediments of Warren volcanic cave, Antarctica, revealed an equal presence of Ascomycota and Basidiomycota taxa, with the complete absence of Chytridiomycota and Mucoromycota representatives (Connell and Staudigel, Reference Connell and Staudigel2013). A high relative content of Malassezia sp. (indicator of anthropic contamination) was found in this Antarctic cave, while completely absent in Scărișoara ice block, indicating a clean sampling strategy and low anthropic pollutants considering that Scărișoara Ice Cave is open for tourism. The absence of Malassezia sp. (Basidiomycota) could also be explained by the obligate commensal or parasitic metabolism of this species due to the lack of most of the genes encoding carbohydrate-degradation enzymes (Rime and others, Reference Rime, Hartmann and Frey2016). Moreover, Arctic cryoconites from Svalbard area (Edwards and others, Reference Edwards2012) were majorly composed of Basidiomycota phylotypes, with only few cultured taxa belonging to Ascomycota and the complete absence of Chytridiomycota species, unlike Scarisoara cave ice. Interestingly, the psychrophilic Rhodotorula and Mrakia (Basidiomycota) species able to degrade high molecular weight PAHs and pectate lyase producers (Margesin and others, Reference Margesin, Fauster and Fonteyne2005; Hesham and others, Reference Hesham2012) were present in both icy environments, while the Ascomycota species Articulospora, Preussia and Pseudeurotium were present only in cryoconites (Edwards and others, Reference Edwards2012). These differences could be associated with the different depositional sources of the Arctic cryoconites and perennial ice block from Scarisoara cave.
The cultured fungal community from Scărișoara perennial ice formed during the last 900 years (Brad and others, Reference Brad2018) revealed the presence of the same four fungal phyla Ascomycota, Basidiomycota, Chytridiomycota and Mucoromycota but with a dissimilar taxa composition and distribution across the ice block at the genera level. Thus, Mrakia stokesii was the dominating species within the cultured cave ice community, present in all analysed strata, while our survey highlighted the occurrence of C. victoriae as the ubiquitously distributed fungal phylotype within the environmental communities across the cave ice block up to 1500 years old strata. This species, also identified in the alpine ice cave (Margesin and others, Reference Margesin, Gander, Zacke, Gounot and Schinner2003), was reported as the predominant Basidiomycota yeast in glacial biomes of both Patagonia (Argentina) and the Svalbard archipelago (Norway) (Garcia and others, Reference Garcia, Zalar, Brizzio, Gunde-Cimerman and Broock2012), also belonging to the most abundant fungal genus identified in soils of the McMurdo Dry Valleys (Connell and Staudigel, Reference Connell and Staudigel2013). Although the cave ice block was dominated by the phylum Ascomycota, this Basidiomycota species appeared to be present in all ice strata. This could be due to specific adaptation mechanisms to harsh environments of Cryptococcus species leading to a high stress tolerance in glacial habitats (low temperatures and low nutrient concentrations) and ability to degrade organic macromolecules by secreting extracellular hydrolytic enzymes (Garcia and others, Reference Garcia, Zalar, Brizzio, Gunde-Cimerman and Broock2012).
Impact of climate and ice geochemistry on the fungal community
The composition of fungal communities entrapped in cave ice layers of different ages appeared to be affected by climate variations during ice deposition, with Ascomycota dominating the ice strata formed during the LIA and DACP and a reduced presence of Mucoromycota phylum, while Basidiomycota was majorly present in 900 years old ice deposited during the warmer and wetter MWP. However, the fungal community composition of the 400 and 1500 years old ice formed during the two cold periods was strongly dissimilar at both class and genus levels, suggesting more complex depositional processes that modelled the fungal community structure within Scărișoara ice block.
Our survey revealed a major representation of plant parasitic fungi and plant/dead wood decomposers belonging to Ascomycota (Schoch and others, Reference Schoch2009) in the 400 (LIA) and 1500 (DACP) years old ice strata formed during the cold intervals. Among these, Phaeosphaeria sp., known as necrotrophic and saprobic species for a wide range of plants (Quaedvlieg and others, Reference Quaedvlieg2013), appeared to be one of the most abundant phylotype of the 1500 years old cave ice layer (Table S4). This OTU was also found in Antarctic soil, confirming the widespread of fungal species in various frozen habitats worldwide. Moreover, ice strata formed during the cold periods had a high relative content of the plant pathogens Mycosphaerella sp. (Crous, Reference Crous2009) and Alternaria sp. (Woudenberg and others, Reference Woudenberg2015), also found in various tree species (beech, Picea abies, and Kunzea ericoides) from cold/alpine and dry habitats (Table S4). During these intervals, the surrounding vegetation of Scărișoara Ice Cave was dominated by spruce (P. abies) forests (Feurdean and others, Reference Feurdean, Perşoiu, Pazdur and Onac2011), that appeared to be a favourable host for parasitic Ascomycota fungi (Jasalavich and others, Reference Jasalavich, Ostrofsky and Jellison2000). This climate driven preferred vegetation was confirmed by the presence of Cadophora sp. (OTU 210) and uncultured fungal taxa isolated from pine and spruce trees (OTU 22) in the LIA-formed ice sample 400-O.
Members of Basidiomycota were highly present in 900-I ice formed during the warmer and wetter MWP. Taxa affiliated to the Microbotryomycetes class were mainly present in the 900 years old clear ice layer, corresponding to a variety of black yeasts belonging to Mrakia sp., C. victoriae and Rhodotorula sp. (Kan and others, Reference Kan2018). These phylotypes, also found in cultured fungal communities from Scărișoara Ice Cave in both 1 and 900 years old ice layers (Brad and others, Reference Brad2018), have been recently identified in 10 000 years old permafrost from Swiss Alps (Frey and others, Reference Frey2016).
Based on these observations, the fungal community from this glacial habitat appeared to constitute a putative reservoir for identification of microbial proxies for climate variations, considering the dominance of Ascomycota and Basidiomycota taxa associated with warmer and colder climatic periods, respectively.
The geochemical composition in different horizons of ice could be associated with climate variations that could affect the ice deposition process. Thus, the cold and dry conditions reduced the hydrological cycle and precipitation rate, increasing the aerosols and dust residence time and allowing their dispersion to great distances. Various element ice contents related to the genesis of the deposited aerosols, where Ca is a well-known ion indicator of terrestrial dust (Zhang and others, Reference Zhang, Yao, An, Tian and Xu2006), high Na and Cl concentrations correspond to sea salt aerosol origin, and high of SO4 and Cl concentrations to a significant volcanic aerosol deposition (Miteva and others, Reference Miteva, Teacher, Sowers and Brenchley2009). Our data indicated a dominant terrestrial dust deposition based on the high concentration of Ca in all ice strata, whereas a small concentration of Na/Cl could lead to a scarce deposition of sea-originating aerosol. The distribution of fungal community across Scarisoara cave ice block showed a high correlation with Ca concentrations, in particular during colder periods, which could result from more cryogenic carbonate formation during prolonged and/or intensive freezing periods (Žák and others, Reference Žák, Onac and Perşoiu2008). Interestingly, the cave ice layers 400-O and 1500-I formed during the cold periods were characterised by neutral pH values, while the ice formed during the MWP was slightly alkaline, suggesting a pH-driven fungal community structure in this glacial habitat. During these colder periods, P. abies forests dominated the cave's surroundings, allegedly inducing a lower soil pH due to the high content of shikimic and quinic acids contained in needles falling on the ground (Lindner and Grill, Reference Lindner and Grill1978). Meanwhile, the fungal diversity of 900-O and 900-I ice layers formed during MWP showed a significantly different response to pH, with a clear clustering of 900-I sample, unlike the bacterial communities from these ice strata that had a similar response to pH (Iţcuş and others, Reference Iţcuş, Pascu, Brad, Perşoiu and Purcarea2016). In addition, the presence in the recently formed cave ice of Hypogymnia species (Table S4) known as organic acid compounds fungal producers (Białońska and Dayan, Reference Białońska and Dayan2005) could be explained by the lower pH values of these ice deposits.
The organic carbon content of ice strata also appeared to play a modelling role of the fungal community in Scărișoara Ice Cave. A relatively low occurrence of Basidiomycota OTUs was found across the Scarisoara ice block (~10% relative abundance) except for the clear ice sample 900-I (90%), characterised by the lowest DOC content and higher pH, suggesting that taxa belonging to this phylum are outcompeted by saprotrophs in the presence of high DOC contents, although the basidiomycetous yeast Cryptococcus sp. was widespread across the ice layers sustained by its cellulolytic and hemicellulolytic activities (Rime and others, Reference Rime, Hartmann and Frey2016), and by a particular cell wall composition enabling this species to withstand a wide pH range (Garcia and others, Reference Garcia, Zalar, Brizzio, Gunde-Cimerman and Broock2012). However, no significant variation of this taxa representation was observed in the 1-S and 1-L samples, suggesting a more complex modelling mechanism of the fungal community in recent ice deposits. Moreover, one of the dominating species of the 900 years old ice (Table S4) was the lichenised Ascomycota Myrothecium sp., representing a potent cellulose decomposer (Haghighi and Shahdoust, Reference Haghighi and Shahdoust2015), in accordance with the low DOC content of this ice layer.
CONCLUSION
This study provided the first high-throughput sequencing across 1500 years old ice revealing a broad fungal diversity in Scărișoara Ice Cave. Various factors such as age of ice, substrate geochemistry and climate patterns during ice formation appeared to influence the diversity and distribution of fungi in the perennial cave ice block. The current data sustain the possibility to identify fungal biomarkers for climate variations in this cave perennial ice, consolidating the relationships between microbial population and environmental characteristics. Further investigations of ice-entrapped microorganisms will extend the assessment of microbiome composition and role in this habitat, improving our understanding on the effect of local climate on the origin and microbial composition at different ice depths. Moreover, novel fungal strains could be further identified and screened for various biotechnological applications using the identified cave ice taxa.
SUPPLEMENTARY MATERIAL
The supplementary material for this article can be found at https://doi.org/10.1017/aog.2019.6
ACKNOWLEDGEMENTS
We thank Christian A. Ciubotărescu, Alexandra Hillebrand-Voiculescu, Traian Brad, Madalina D. Pascu, Bogdan P. Onac, Carmen Bădăluță and Viorica Nagavciuc for assistance with ice sampling, and Pablo Arán Sekul for bioinformatics support. This work was financially supported by the H2020 EraNet-LAC ELAC2014/DCC0178 Joint Program and UEFISCDI grants PN-II-ID-PCE-2011-3-0742 and PN-III-P1-1.1-TE-2016-2210. This project has received funding from the European Union's Horizon 2020 research and innovation programme under the Marie Skłodowska-Curie grant agreement No. 675546.
AUTHOR CONTRIBUTIONS
AM and CP wrote the manuscript, with important contribution from AP. CI and AP participated in ice sampling; CI conducted the sample filtration and DNA extraction; BF performed the bioinformatics analyses; BF and JD carried out the statistical analyses; PL performed BLAST sequence analysis; CM conducted the geochemical analyses; AP contributed to 14C radiocarbon ice dating; CP performed the experimental design and coordinated the project. All of the authors contributed to data interpretation and revision of the manuscript and approved its final version.










