Introduction
Rationale
Mastitis is one of the most costly diseases of dairy cattle (Halasa et al., Reference Halasa, Huijps, Østeräs and Hogeveen2007). It is painful, can result in premature culling, reduced milk production, and decreased fecundity at the cow level, and is often accompanied by herd-level consequences such as poor milk quality and increased risk of antimicrobial residues in marketed milk. In the United States, treatment for clinical mastitis represents the most common indication for antimicrobial use in adult dairy cattle, with 16.4% of cows reported as treated in 2007, with cephalosporins the most commonly selected drug class (United States Department of Agriculture, 2008). While the bacterial etiology varies, a significant proportion of these cases benefit from prompt administration of an effective antimicrobial, with or without other therapy. Dairy farmers and veterinarians have a considerable number of antimicrobial treatments available, including products of greater importance to human medicine. Veterinarians need information about the relative efficacy among products to facilitate their choices and, where possible, select efficacious products with the lowest human medical importance.
For many clinical mastitis treatments, comparative efficacy estimates are available for only one or two antimicrobial products. Ideally, producers and veterinarians would have comparative efficacy of all possible treatment options, to allow the inclusion of relative efficacy with other treatment decision parameters such as cost, convenience, and importance to human medicine. Traditionally, information about efficacy would be obtained from randomized controlled trials; however, with so many treatment options for clinical mastitis, a trial that concurrently includes all possible treatment options is not feasible. A robust alternative is to conduct a network meta-analysis that combines all of the information from multiple trials and enables accurate and valid comparisons to be made for all available treatments. The statistical methods for this approach are well established and have been used extensively in human health (Dias et al., Reference Dias, Ades, Welton, Jansen and Sutton2018) and are beginning to become more common in animal health research (O'Connor et al., Reference O'Connor, Coetzee, da Silva and Wang2013, Reference O'Connor, Yuan, Coetzee, da Silva and Wang2016). Network meta-analysis provides a method of assessing relative efficacy among many treatments by use of direct (studies which compare given treatments) and indirect (studies which share common comparators) evidence, and is a commonly used approach in human medicine (Caldwell et al., Reference Caldwell, Ades and Higgins2005; Cipriani et al., Reference Cipriani, Higgins, Geddes and Salanti2013).
Establishing the relative efficacy of antimicrobial therapies for clinical mastitis will serve to improve decision makers' ability to engage in effective stewardship of antimicrobials by avoiding unnecessary use of ineffective therapies, and by allowing the selection of antimicrobials of less importance to human health, if an equally efficacious antimicrobial is available.
This systematic review was conducted based on the guidance from the Cochrane Collaboration (Higgins and Green, Reference Higgins, Thomas, Chandler, Cumpston, Li, Page and Welch2011) and recommendations for conducting systematic reviews in animal agriculture and veterinary medicine (O'Connor et al., Reference O'Connor, Anderson, Goodell and Sargeant2014a, Reference O'Connor, Sargeant and Wang2014b; Sargeant and O'Connor, Reference Sargeant and O'Connor2014a, Reference Sargeant and O'Connor2014b; Sargeant et al., Reference Sargeant, Kelton and O'Connor2014a, Reference Sargeant, Kelton and O'Connor2014b). It is reported using the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) Extension Statement for Reporting of Systematic Reviews Incorporating Network Meta-analyses of Health Care Interventions (PRISMA-NMA) (Hutton et al., Reference Hutton, Salanti, Caldwell, Chaimani, Schmid, Cameron, Ioannidis, Straus, Thorlund, Jansen, Mulrow, Catala-Lopez, Gotzsche, Dickersin, Boutron, Altman and Moher2015).
Objectives
The objective of this review was to conduct a systematic review and network meta-analyses to summarize the results of clinical trials conducted to evaluate the efficacy of antimicrobials for the treatment of clinical mastitis in dairy cattle, for both bacteriologic and clinical cure.
Methods
Protocol
A review protocol, reported in accordance with PRISMA-P guidelines (Moher et al., Reference Moher, Shamseer, Clarke, Ghersi, Liberati, Petticrew, Shekelle, Stewart and PRISMA-P Group2015), was published to the University of Guelph's institutional repository (https://atrium.lib.uoguelph.ca/xmlui/handle/10214/10046) on 2 January 2018. The protocol is also available through Systematic Reviews for Animals and Food (SYREAF) (http://www.syreaf.org/contact/).
Eligibility criteria
English-language, primary research studies were eligible for inclusion. Studies must have been conducted in lactating dairy-breed cattle with naturally occurring clinical mastitis. At least one treatment arm must have included an antimicrobial treatment (given after diagnosis of clinical mastitis) by any route, compared to no treatment, placebo, or an alternative treatment (such as another antimicrobial). Table 1 further describes the treatment groups. Interventions of long-acting single-dose antimicrobials designed to be given to cows at dry-off were ineligible. Eligible outcomes were bacteriologic cure or clinical cure (both as defined by the authors). Controlled trials with natural disease exposure were the only eligible study design.
Table 1. Description of treatment groups as labeled in subsequent figures and tables

Search
Sources
Electronic searches were completed using CABI (via CAB Direct, 1973 to current), MEDLINE (via Ovid, 1950 to current), Agricola (via ProQuest, 1970 to current), and Toxline (via ProQuest, 1840 to current) databases. ProQuest Theses and Dissertations (via ProQuest Central, 1997 to current) were searched. A single reviewer hand-searched the table of contents of the National Mastitis Council (NMC) and American Association of Bovine Practitioners (AABP) conference proceedings from 1997 to 2018, as well as the Bovine Practitioner journal. The Food and Drug Administration (FDA) website containing the Freedom of Information New Animal Drug Approvals (NADA) summaries were also searched by examining all available online summaries.
Search strategy
The search strategy was initially developed for CABI (via Cab Direct) and consisted of three concepts: dairy cattle (including breed terms and mastitis terms), antimicrobial treatment, and outcome terms (clinical or bacteriologic cure). The full search strategy is listed in Table 2. Database searches were conducted on 2 February 2018 (CABI) and 12 February 2018 (Medline, Agricola, Toxline, and ProQuest Theses and Dissertations). Results from the searches were uploaded to EndNoteX7 (Clarivate Analytics, Philadelphia, PA, USA), and then exported and uploaded to DistillerSR (Evidence Partners Inc., Ottawa, ON, USA). Duplicate records were documented and removed through both EndNoteX7 and DistillerSR. If the same study appeared as both a conference proceeding and a full publication, the conference proceeding abstract was removed. Data only available as a conference proceeding were eligible if the full text was >500 words, to allow sufficient detail for data extraction and risk of bias assessment.
Table 2. Full electronic search strategy used to identify studies of the efficacy of antimicrobial therapy for treatment of clinical mastitis in lactating dairy cattle in CABI (via CAB Direct Web) conducted on 2 February 2018

Study selection
Distiller SR was used for relevance screening and data extraction. All screening was conducted independently in duplicate by CBW and KJC or JMS. The first 250 titles and abstracts were used to pre-test the questions to ensure clarity and consistency of question application. Initially, title and abstract were screened for relevance using the following questions:
(1) Does the title and/or abstract describe a controlled trial? YES (neutral), NO (exclude), UNCLEAR (neutral);
(2) Does the title and/or abstract describe a study of naturally occurring clinical mastitis in lactating dairy cows? YES (neutral), NO (exclude), UNCLEAR (neutral);
(3) Does the title and/or abstract describe one or more intervention groups of an antimicrobial therapy, as compared to either another antimicrobial therapy, a placebo, or no treatment? YES (neutral), NO (exclude), UNCLEAR (neutral);
(4) Is at least one of the mastitis treatments described above NOT a long-acting, single treatment therapy designed to be given to cows at dry-off? YES (include for full-text screening), NO (exclude), UNCLEAR (include for full-text screening)
Citations were excluded if both reviewers agreed the study did not meet one or more of the descriptions by answering ‘No’ to any of the previous questions. Disagreements were resolved by consensus with mediation by JMS or DFK if an agreement could not be reached. Full-text screening was then conducted by KJC and CBW independently in duplicate for all studies passing the primary round, using the first 10 citations as a pre-test. This screening level used the previous four questions with only ‘YES’ (neutral) or ‘NO’ (exclude) options, and additionally:
(1) Is the full text available in English? YES (neutral), NO (exclude);
(2) Is at least one of the mastitis treatments described above given after the diagnosis of clinical mastitis (i.e. not used for prevention)? YES (neutral), NO (exclude);
(3) Does the study describe an outcome of either bacteriologic or clinical cure? YES (include for data extraction), NO (exclude)
Citations were excluded if both reviewers agreed the study did not meet the inclusion criteria by answering ‘No’ to any of the above questions. Agreement for exclusion at the full-text screening was at the question level, with conflicts resolved by consensus with mediation by JMS if an agreement could not be reached.
Data collection
Data from citations meeting the full-text inclusion criteria were extracted independently in duplicate by CBW and KJC using a standardized form, which was pre-tested on the first five articles to ensure clarity and consistency of application. Discrepancies in data extraction were resolved by consensus, with mediation by JMS if an agreement could not be reached. A PDF version of the data extraction tool is available as Supplementary File S1.
Data items
Study characteristics
Study-level data included study location, year of conduct, cattle breed, farm type (commercial or research), number of farms enrolled, cow-level inclusion criteria (lactation, number of previous clinical mastitis events, days since last event, other (specify)), who determined the diagnosis (producer, veterinarian, or researcher), how severity was scored, who collected the pre-treatment sample (producer, veterinarian, or researcher), who was responsible for treatment (producer, veterinarian, or researcher), and when treatment was administered relative to diagnosis.
Interventions and comparators
Details on the interventions recorded included the type of antimicrobial drug, route, frequency, duration, concurrent therapy (including if additional supportive and/or anti-inflammatory treatment was allowed), and level of treatment allocation. While results of all comparisons in the network were included in the analysis, only treatment arms with an intramammary treatment labeled in North America for use in lactating dairy cattle are presented with relative efficacy rankings (i.e. other therapies were not ranked, but information captured on these comparator arms provided evidence to the network).
Eligible outcomes
Clinical cure and bacteriologic cure(s) (by pathogen or species group) were extracted for all trials with data presented numerically. Multiple definitions of a single outcome were extracted if reported (e.g. bacteriologic cure at 7 days, and at 21 days) but only one definition (longest cure length) was included in the meta-analysis.
For trials reporting clinical cure, the duration of time between treatment and measurement of the outcome was recorded, as well as the definition of cure and level of analysis (quarter or cow). For bacteriologic cure, how cure was defined, details on how bacteriology was performed, the species or species groups reported, when cure was assessed relative to end of treatment, and level of analysis were recorded. An a priori decision was made with respect to combining bacteriologic diagnostic information to group pathogen species that were likely to have a similar response to treatment. Misclassification of Enterococcus species as Streptococcus prior to use of MALDI-TOF mass spectrometry was a concern, and data therefore were not combined for general Strep. species as this likely would have included misclassified Enterococcus. Staphylococcus aureus were considered separately from coagulase-negative Staph species or non-aureus Staph species, which were grouped. Streptoccus aglactiae, S. uberis, and S. dysgalactiae were considered unique diagnoses, while Enterococcus species were considered a unique group. Escherichia coli and Klebsiella species were considered as separate diagnoses.
For eligible outcomes, the prioritized outcome metric was treatment arm-level data (raw data), as this was thought to be the most prevalent form of data in the literature. Data not presented as numbers (i.e. data points in graphs) were not extracted.
Geometry of the network
The geometry of the network was visually examined to determine if some pairwise comparisons dominated and to determine the network structure, and if there were intervention comparisons that were not linked to the network (i.e. did not have an intervention in common with one or more other published trials).
Risk of bias in individual trials
The risk of bias was assessed by outcome for each outcome extracted, using the Cochrane Risk of Bias 2.0 tool (Higgins et al., Reference Higgins, Sterne, Savovic, Page, Hróbjartsson and Boutron2016), with signaling questions modified to be specific to the topic of the review (available as Supplementary File S2). This tool assesses the potential for bias arising from five areas or domains: bias arising from the randomization process, bias due to deviations from intended interventions, bias due to missing outcome data, bias in the measurement of the outcome, and bias in the selection of the reported results. An additional potential response was provided for the question on random allocation sequence, for trials using the word ‘random’ to describe the allocation sequence but not providing details on the method used to generate the random sequence.
Risk of bias was assessed independently in duplicate, with disagreement resolved by consensus and mediation by JMS or CBW if needed. Risk of bias is presented by outcome, then by domain of bias, in order to identify which areas of bias had specific deficiencies.
Summary measures
After extracting the outcomes, the analysis was conducted on the log OR. For presentation purposes, the log odds ratio was back transformed to the risk ratio (RR) using the baseline risk from the model data. The posterior mean and standard deviation of the baseline risk mean were −0.018 and 0.8697. The posterior mean and standard deviation of the baseline risk standard deviation were 1.1594 and 0.6285. When trials had zero cells for some data points, and the odds ratio could not be calculated, the trial results could not be included in the analyses.
Network meta-analysis
Planned method of statistical analysis
A network meta-analysis was planned for the outcomes of clinical and bacteriologic cure(s). The method has been previously described in detail elsewhere (Dias et al., Reference Dias, Welton, Caldwell and Ades2010). Briefly
 $$r_{\,jk}\,\sim \,{\rm Bin}\,\lpar p_{\,jk}\comma \;n_{\,jk}\rpar \,\comma \;\,\theta _{\,jk}\,= \,{\rm logit}\,\lpar p_{\,jk}\rpar $$
$$r_{\,jk}\,\sim \,{\rm Bin}\,\lpar p_{\,jk}\comma \;n_{\,jk}\rpar \,\comma \;\,\theta _{\,jk}\,= \,{\rm logit}\,\lpar p_{\,jk}\rpar $$and
 $$\theta _{\,jk}\,= \,\lcub \mu _{\,jb}\comma \;\,{\rm if}\,k\,= \,b\semicolon \;\,b\,= \,A\comma \;B\comma \;C\comma \;\ldots \mu _{\,jk}\,+ \,\delta _{\,jk}\,\comma \;\,{\rm if}\,k\,\gt \,b\semicolon \;b\,= \,A\comma \;B\comma \;C\,\ldots $$
$$\theta _{\,jk}\,= \,\lcub \mu _{\,jb}\comma \;\,{\rm if}\,k\,= \,b\semicolon \;\,b\,= \,A\comma \;B\comma \;C\comma \;\ldots \mu _{\,jk}\,+ \,\delta _{\,jk}\,\comma \;\,{\rm if}\,k\,\gt \,b\semicolon \;b\,= \,A\comma \;B\comma \;C\,\ldots $$where pjk is the probability of the event in trial j under treatment k and δjbk is the trial-specific log odds ratio of treatment k relative to the corresponding baseline treatment b in trial j. The trial-specific treatment effects are distributed as:
 $$\delta _{\,jbk}\,\sim \,N\lpar d_{bk}\comma \;\sigma ^2_{bk} \rpar \comma \;$$
$$\delta _{\,jbk}\,\sim \,N\lpar d_{bk}\comma \;\sigma ^2_{bk} \rpar \comma \;$$with priors
 $$d_{bk}\,\sim \,N\,\lpar 0\comma \;10000\rpar $$
$$d_{bk}\,\sim \,N\,\lpar 0\comma \;10000\rpar $$and under the homogeneous variance assumption that σ 2bk = σ 2 where σ ~ U(0,5)
For multi-arm trials, we assumed the co-variance between δjAB and δjAC was σ 2/2 (Higgins and Whitehead, Reference Higgins and Whitehead1996; Lu and Ades, Reference Lu and Ades2004).
Selection of prior distributions in Bayesian analysis
The prior distributions were originally based on the approach reported previously (Dias et al., Reference Dias, Welton, Sutton and Ades2011). For the model, we assessed σ ~ U (0,2) and σ ~ U (0,5). As the analysis suggested σ ~ U (0,5) was preferred, we retained this prior in the model.
Implementation and output
All posterior samples were generated using Markov Chain Monte Carlo (MCMC) simulation implemented using Just Another Gibbs Sampler (JAGS) software (version 3.4.0) (Plummer, Reference Plummer2015). All statistical analyses were performed using R software (version 3.2.1) (R Core, 2015) in a Linux system. The model was fit by calling JAGS from R through the RJAGS package (Plummer, Reference Plummer2015). Three chains were simulated and the convergence was assessed using Gelman–Rubin diagnostics. A total of 5000 ‘burn-in’ iterations were discarded, and based the inferences on a further 10,000 iterations. The model output included all possible pairwise comparisons using log odds ratios (for inconsistency assessment), risk ratios (used for comparative efficacy reporting), the rankings (for comparative efficacy reporting), and the probability of being the worst treatment option (for comparative efficacy reporting).
Assessment of model fit
The fit of the model was assessed based on the log odds ratio, by examining the residual deviance between the predicted values from the mixed-treatment comparison model and the observed value for each study (Dias et al., Reference Dias, Welton, Caldwell and Ades2010).
Assessment of inconsistency
Inconsistency was assessed by examining the estimates of treatment effects obtained from direct, combined, and indirect evidence for all pairwise comparisons, using the method described by Dias et al. (Reference Dias, Welton, Caldwell and Ades2010). Means and standard deviations of log odds ratio of treatment effects were calculated using direct (head-to-head) evidence only, indirect evidence only, and the combined evidence. We compared the estimates from the direct and indirect models and considered the standard deviation of each estimate, rather than relying on the P-values.
Risk of bias in the overall network
Risk of bias in the overall network of evidence was assessed using the Confidence In Network Meta-Analysis (CINeMA) platform (http://cinema.ispm.ch), which uses a frequentist approach through the ‘metafor’ package (Viechtbauer, Reference Viechtbauer2010) to determine the basis for the contribution matrix for the risk of bias. CINeMA evaluates within-study bias, across-studies bias, indirectness, imprecision, heterogeneity, and incoherence. As opposed to presenting an overall assessment of bias and of indirectness, we reported the contribution of studies based on the approach to allocation to groups and blinding, as there is evidence in animal health that failure to include these design elements is associated with exaggerated treatment effects (Wellman and O'Connor, Reference Wellman and O'Connor2007; Sargeant et al., Reference Sargeant, Elgie, Valcour, Saint-Onge, Thompson, Marcynuk and Snedeker2009a, Reference Sargeant, Saint-Onge, Valcour, Thompson, Elgie, Snedeker and Marcynuk2009b). Risk of bias due to randomization was assessed as ‘low’ if the authors reported randomization and details of the method used to generate the sequence; ‘some concerns’ if random allocation was reported but no details on how the random sequence was generated were reported; and ‘high’ if no information on allocation was provided or if a non-random method was used. Risk of bias due to blinding was assessed as ‘low’ if both caregivers and outcome assessors were blind to treatment group, ‘unclear’ if caregivers or outcome assessors were blinded, but not both, and ‘high’ if neither caregivers nor outcome assessors were blinded.
Indirectness (how closely the populations studied resemble the target populations for the intervention) was not considered to be an issue due to the eligibility criteria for the review, and therefore the risk of bias was considered ‘low’ for all studies. Across-study bias was not assessed as there are no well-developed methods to determine this in network meta-analysis, and with a small number of pairwise comparisons, the assessment of small study effect would not be informative (Sterne et al., Reference Sterne, Gavaghan and Egger2000). Bias due to imprecision was assessed using 0.8 and 1.25 as the clinically important odds ratios. Similarly, odds ratios of 0.8 and 1.25 were used to assess heterogeneity. Incoherence (inconsistency) analysis was not reported from CINeMA, as this was conducted based on the Bayesian analysis described elsewhere in this paper.
Results
Study selection
The literature search was conducted through the University of Guelph (Medline, Agricola, and Toxline) and Iowa State University (CAB Direct). Results of the search and flow of studies through the screening process are shown in Fig. 1, including reasons for full-text exclusions. The search was validated through the identification of 10 relevant studies pre-selected by DFK that were cross-checked for inclusion.

Fig. 1. Preferred Reporting Items for Systematic Review and Meta-analysis (PRISMA) study flow diagram (Moher et al., Reference Moher, Shamseer, Clarke, Ghersi, Liberati, Petticrew, Shekelle, Stewart and PRISMA-P Group2015).
From an initial search, 14,775 citations were screened by title and abstract, 148 of which proceeded to review of full text. Of these, 54 articles were eligible for data extraction, and they contained 54 unique trials. From these 54 trials, 7 did not have usable data (e.g. data not presented, no variance measure provided for continuous outcomes, or data were presented in graphs or figures only, and so 44 trials had data extracted on one or both outcomes. Clinical cure was reported in 33 trials, and 31 reported bacteriologic cure.
Study characteristics
The 44 trials with data extracted, these were conducted in 20 different countries, most frequently the United States (8/44), Canada (4/44), New Zealand (4/44), and the United Kingdom (4/44). The country of conduct was not reported in 10 (23%) trials. Trial setting was most commonly reported as a commercial dairy or dairies (35/44; 80%), with two trials conducted at research dairies and seven not reporting the trial setting. Twenty-three (52%) trials reported the year of conduct, with six conducted since 2009 (14%), nine (20%) between 2000 and 2009, six (14%) from 1990 to 1999, and two (5%) prior to 1990. Breed was reported in 16 (36%) trials, with Holstein/Friesian being most commonly reported, although trials in other breeds were also identified. Six trials were conducted in a single herd (14%), and the number of herds ranged from 1 to 276. The number of herds used was not reported in 10 trials (23%).
Further details on the 24 trials included in the network meta-analysis of clinical cure are presented in Supplementary File S3, including cow-level eligibility and clinical cure definition.
Outcomes
If outcomes were reported for multiple time points, only the last measure was included for analysis. Thirty-three trials reported clinical cure, and 31 reported bacteriologic cure.
Bacteriologic cure
Although 31 trials examined bacteriologic cure, as not all pathogens were represented in all trials, a smaller number of trials reported bacterial cure risks for each species groups. As well, some trials reported all bacteriologic cures combined together as a single outcome, which was not considered for further analysis. Staphylococcus aureus cure was reported by 22 trials with cure risk reported, 17 trials reported E. coli cure, 15 reported S. uberis cure, 11 reported coagulase-negative Staphylococcus species cure, 11 reported S. dysgalactiae cure, seven reported Streptococcus aglactiae cure, five reported Klebsiella cure, and two reported Enterococcus cure. Bacteriologic cure was measured at the quarter level for all trials. Relative to the last day of treatment, the last assessment of cure ranged from 0 to 30 days.
Clinical cure
Of the 33 trials examining clinical cure, a definition of clinical cure was provided for the majority (28/33). For the last assessment of cure, time from last treatment ranged from 0 to 100 days. For the 24 trials included in the meta-analysis, 21 provided a definition of clinical cure, and assessment of cure was last performed at <7 days in five trials, 7–14 days in two trials, 15–21 days in six trials, 22–28 days in seven trials, and 29–35 days in three trials (days relative to the last treatment administration).
Risk of bias within studies by outcome
Clinical cure
The results of the risk of bias assessment for the 33 trials reporting clinical cure are presented in Fig. 2, showing the risk of bias in the five evaluated domains: bias arising from the randomization process, bias due to deviations from intended interventions, bias due to missing outcome data, bias in the measurement of the outcome, and bias in the selection of the reported results.

Fig. 2. (a) Risk of bias by domain for trials reporting clinical cure of clinical mastitis following therapy that were included in the final network meta-analysis (n = 24). Risk of bias was assessed according to the Revised Cochrane risk-of-bias tool for randomized trials (RoB2) (Higgins et al., Reference Higgins, Sterne, Savovic, Page, Hróbjartsson and Boutron2016). (b) Risk of bias by domain for trials reporting bacteriologic cure of clinical mastitis following therapy (n = 31). Risk of bias was assessed according to the Revised Cochrane risk-of-bias tool for randomized trials (RoB2) (Higgins et al., Reference Higgins, Sterne, Savovic, Page, Hróbjartsson and Boutron2016).
For the risk of bias arising from the randomization process, the majority of trials were assessed as ‘some concerns’ for this domain (27/33), which was primarily driven by a lack of reporting. Ten trials used the word ‘random’ to describe allocation, but did not provide details on how the random sequence was generated, and 28 did not report information regarding whether the allocation sequence was concealed prior to enrollment of animals.
The risk of bias due to deviations from intended interventions was assessed as ‘some concerns’ in the majority of trials (26/33). This was driven primarily through lack of reporting if caregivers were or were not blinded. Treatments were commonly co-mingled in an environmental group, where differential care would be unlikely, and therefore trials not reporting this feature were assessed as ‘some concerns’ and not as ‘high risk’.
The risk of bias due to missing outcome data was assessed as low in 14/33 trials, as a result of either having outcome data available for all, or nearly all, animals which were assigned treatments, or because the proportions of missing data and reasons for missing data were similar across intervention groups. Similar to other domains, lack of reporting drove the assessment of ‘some concerns’ for a majority of trials in this domain, as 13 did not provide information about the number of animals analyzed (e.g. percentage cure was reported as opposed to raw numbers), or no information was provided for why outcome data were missing.
The risk of bias due to measurement of the outcome was assessed as ‘high’ for 28/33 trials, and ‘low’ for the remaining five trials. ‘Low’ risk of bias was assigned if outcome assessors were blinded to treatment group. As clinical cure is a subjective measure, a lack of information on blinding or reporting that blinding was not used, resulted in a ‘high’ risk of bias assessment. Eight trials reported that blinding was not used, and 20 did not provide any information on blinding.
For the risk of bias arising from the selection of the reported results, information regarding a priori intentions of outcome measurements and analyses were not available for any trials; this domain generally requires the examination of a trial protocol or statistical analysis plan documented before the trial if there are multiple ways an outcome could be measured or analyzed. As a result, all trials were assessed as ‘some concerns’ in this domain.
Bacteriologic cure
Risk of bias for each domain, and reasons for determination, were similar for trials reporting bacteriologic cure (n = 31) compared to those reporting clinical cure. For trials reporting both outcomes, the risk of bias resulting from the randomization process or deviations from intended interventions were the same. The risk of bias due to missing outcome data was the same, with a number of trials failing to report the number of study units analyzed or why outcome data were missing. The risk of bias due to the measurement of the outcome was assessed as ‘low’ for all trials, as the assessment of the outcome was unlikely to be influenced by knowledge of the intervention. As a result, regardless of blinding status, all trials fell into the ‘low-risk’ category. As there were multiple ways the outcome could be measured or analyzed, and no trials gave information regarding an a priori trial protocol or analysis plan, all were assessed as ‘some concerns’ for this domain.
Quantitative summary
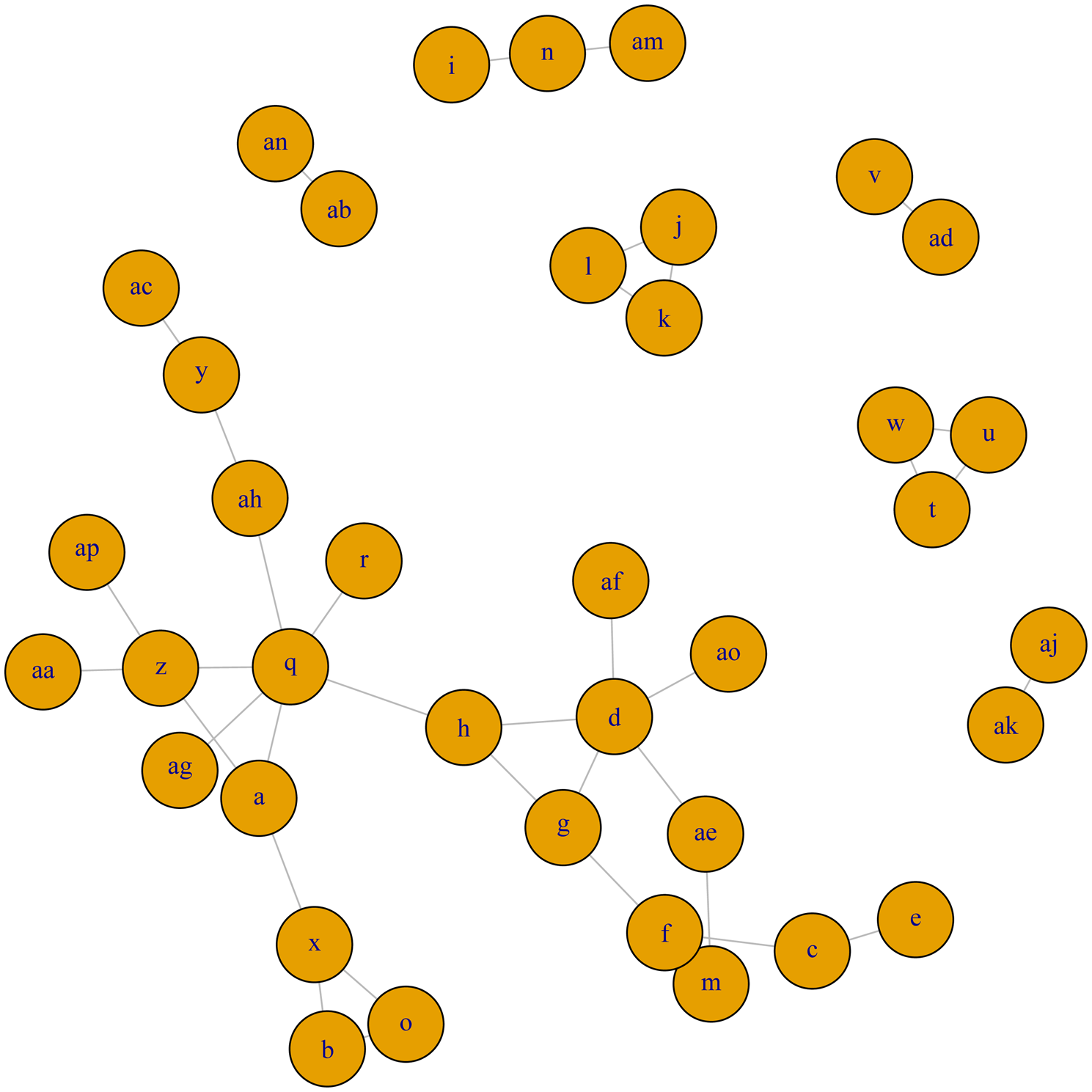
A network meta-analysis was conducted for trials examining clinical cure following antimicrobial therapy for clinical mastitis in lactating dairy cows. Although network meta-analysis was planned for major bacteriologic categories, none of the networks were considered large and diverse enough to conduct a meaningful network meta-analysis. For coagulase-negative Staphylococcus species (Fig. 3a), E. coli (Fig. 3b), S. aureus (Fig. 3c), S. dysgalactiae (Fig. 3d), and S. uberis (Fig. 3e), the networks of evidence were very disconnected, i.e. common treatment arms did not exist between many of the studied treatments. As a result, it was not possible to provide relative comparisons of treatments which did not share a common comparator, nor was additional strength provided by the use of indirect estimates, as compared to the larger, more connected network of trials for the clinical cure outcome (Fig. 4). For S. aglactiae, Klebsiella, and Enterococcus, too few trials were found to inform the composition of a treatment network.

Fig. 3. (a) Full network plot for the examination of relative efficacy of lactating cow antimicrobial therapy on bacteriologic cure of coagulase-negative Staphylococcus species. Green circles indicate intramammary treatments of antimicrobials in an OIE category which contains a currently labeled product available in North America. Red circles indicate a therapy which is a route other than intramammary, or with a product in an OIE category which does not have a currently labeled intramammary producer in North America. Full treatment arm descriptions are found in Table 1. (b) Full network plot for the examination of relative efficacy of lactating cow antimicrobial therapy on bacteriologic cure of Escherichia coli. Green circles indicate intramammary treatments of antimicrobials in an OIE category which contains a currently labeled product available in North America. Red circles indicate a therapy which is a route other than intramammary, or with a product in an OIE category which does not have a currently labeled intramammary product in North America. Full treatment arm descriptions are found in Table 1. (c) Full network plot for the examination of relative efficacy of lactating cow antimicrobial therapy on bacteriologic cure of Staphylococcus aureus. Green circles indicate intramammary treatments of antimicrobials in an OIE category which contains a currently labeled product available in North America. Red circles indicate a therapy which is a route other than intramammary, or with a product in an OIE category which does not have a currently labeled intramammary product in North America. Full treatment arm descriptions are found in Table 1. (d) Full network plot for the examination of relative efficacy of lactating cow antimicrobial therapy on bacteriologic cure of Streptococcus dysgalactiae. Green circles indicate intramammary treatments of antimicrobials in an OIE category which contains a currently labeled product available in North America. Red circles indicate a therapy which is a route other than intramammary, or with a product in an OIE category which does not have a currently labeled intramammary product in North America. Full treatment arm descriptions are found in Table 1. (e) Full network plot for the examination of relative efficacy of lactating cow antimicrobial therapy on bacteriologic cure of Streptococcus uberis. Green circles indicate intramammary treatments of antimicrobials in an OIE category which contains a currently labeled product available in North America. Red circles indicate a therapy which is a route other than intramammary, or with a product in an OIE category which does not have a currently labeled intramammary producer in North America. Full treatment arm descriptions are found in Table 1.
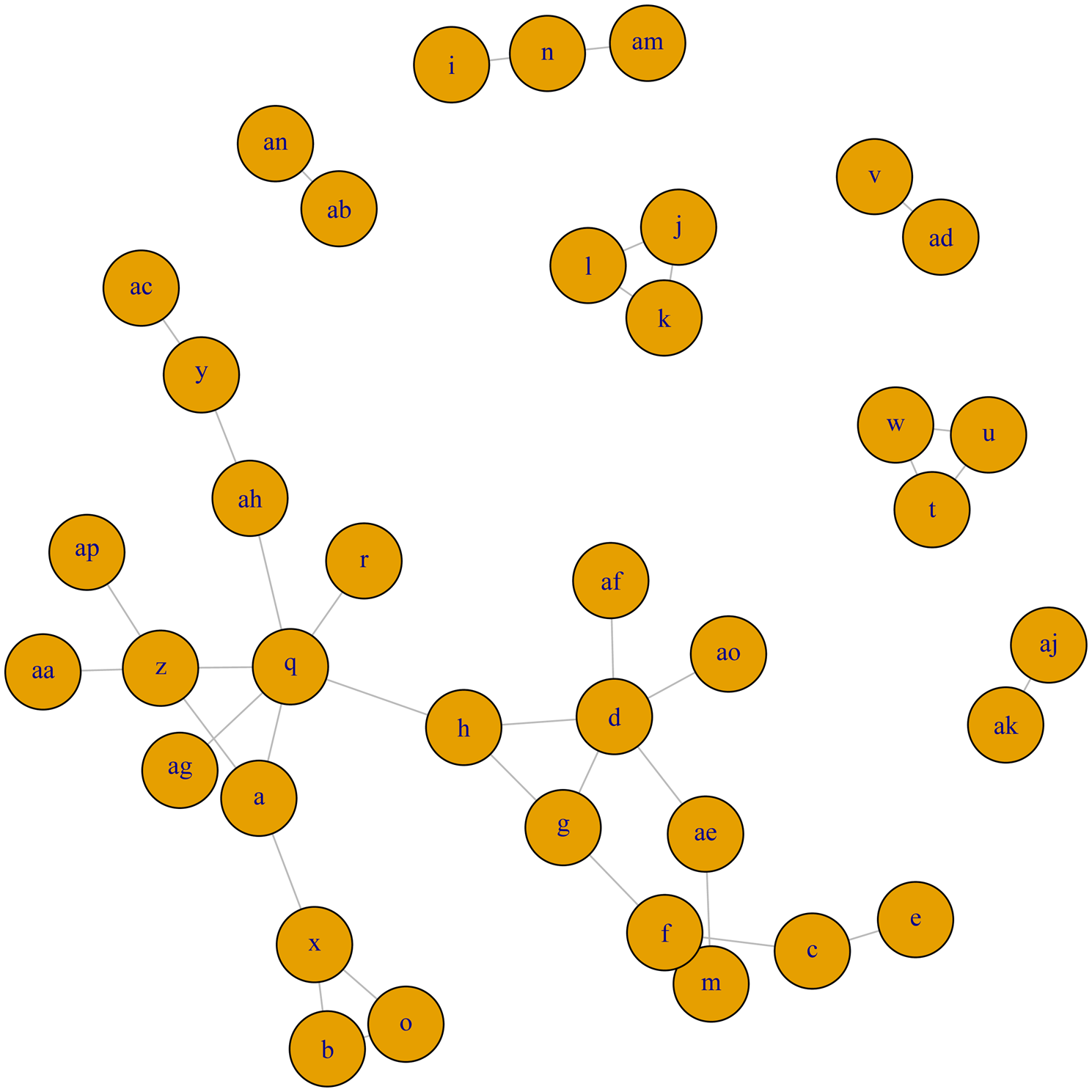
Network meta-analysis – clinical cure
The full network plot for clinical cure is shown in Fig. 4. Fifteen treatment arms were not connected to the larger network (and, were each only connected to one or two other arms), and so could not be included in the meta-analysis. The network of evidence used in meta-analysis is shown in Fig. 5, and represents 51 treatment arms from 24 trials, including 21 two-arm trials and three three-arm trials.

Fig. 5. Treatment arm network for the examination of relative efficacy of lactating cow antimicrobial therapy for clinical mastitis on clinical cure rate. The size of the circle indicates the relative number of arms and the width of the lines indicates the relative number of direct comparisons. Full treatment arm descriptions are found in Table 1. Treatment arms c, d, e, m, q, s, x, z, ae, and ap are intramammary therapies from categories with current labeling for lactating cow therapy in North America, treatment arm f is non-treated control or placebo group.
Assessment of consistency
The consistency assessment relates only to outcome (clinical cure) for which a network meta-analysis was conducted, and is shown in Table 3. The means and standard deviations of log odds ratio of treatment effects are shown using direct (head-to-head) evidence only, indirect evidence only, and the combined evidence. The inconsistency estimate and standard deviation are presented. The approach used to assess inconsistency was to evaluate the direction of the estimates and determine if they were indicative of different effects combined with an evaluation of the width of the credible intervals. Using this approach, there was one of 25 comparisons which had a concern regarding the inconsistency estimate: treatment h (intramammary quinolone) compared to treatment g (parenteral quinolone).
Table 3. Direct (dir) and indirect (rest) comparisons for the consistency assumption

The inconsistency estimate (ωXY) and standard deviation (SDωXY) are shown. Posterior means (d) and standard deviation (SD) of the log odds ratio of intervention effects calculated for direct (head-to-head) evidence only (dir), indirect evidence only (rest) and a combination of all evidence (MTC). The first treatment listed is the referent (denominator) and the second listed is the comparator (numerator).
The contribution of trials to estimates based on the randomization status of the study is presented in Fig. 6, and contribution of studies to estimates based on blinding is presented in Fig. 7. Most pairwise comparisons included a roughly equal contribution from trials which randomly allocated to treatment and provided evidence of random sequence generation (green), and those which did not report random allocation or reported a non-random method (red) (Fig. 6). A smaller contribution came from trials which described random allocation with no supporting evidence (yellow). For contribution of trials to estimates based on blinding (Fig. 7), in most pairwise comparisons, there was only a very small (or no) contribution from studies reporting blinding of both (green) or either (yellow) caregivers or outcome assessors. Table 4 further summarizes the risk of bias conclusion for each pairwise comparison for randomization, blinding, imprecision, and heterogeneity.

Fig. 6. Contribution of studies to the point estimates based on the description of allocation approach for studies contributing to the network meta-analysis examining the relative efficacy of intramammary lactating cow therapy on the risk of clinical cure (n = 24). Green indicates studies that randomly allocated to treatment and provided evidence of random sequence generation, yellow indicates studies that reported random allocation but did not provide supporting evidence, and red indicates studies that did not report allocation approach or reported a non-random method. White vertical lines indicate the percentage contribution of separate studies.

Fig. 7. Contribution of studies to the point estimates based on the description of blinding for studies contributing to the network meta-analysis examining the relative efficacy of intramammary lactating cow therapy on the risk of clinical cure (n = 24). Green indicates studies that reported both caregivers and outcome assessors were blinded to treatments, yellow indicates studies that reported caregivers or outcome assessors were blinded to treatment (but not both), and red indicates studies where blinding was not used, or not reported, for both caregivers and outcome assessors. White vertical lines indicate the percentage contribution of separate studies.
Table 4. Summary of the overall quality of evidence of the network of studies examining the efficacy of intramammary lactating therapy on clinical cure, using the Confidence In Network Meta-Analysis (CINeMA) platform (http://cinema.ispm.ch), with a modified approach, to determine the risk of bias due to approach to randomization, blinding, imprecision, and heterogeneity

Imprecision and heterogeneity were determined using a clinically important odds ratio of 0.8.
Rankings and distribution probability of clinical cure
The rankings and probability distributions are only shown for intramammary antimicrobials belonging to a category where there is a currently labeled product in North America. Risk ratios from the network meta-analysis comparing these treatments are shown in Table 5. The risk ratio is the risk of the event (clinical cure) in the column heading (the treatment arm included as the numerator in the risk ratio), divided by the risk of the event in the row header (the treatment arm included as the denominator in the risk ratio). For example, the estimated probability of clinical cure for f (non-treated control) is 0.69 times the probability of cure for cows treated with intramammary penicillin (treatment z). The corresponding confidence interval is located in the lower left-hand section of the table, with rows and columns reversed to those used for the risk ratio (95% CI 0.30–1.59). Although almost all products were numerically better than non-treated controls, all credibility intervals overlapped a value of one, as do pairwise comparisons between products, meaning results are consistent with no effect. The distribution of the probability of clinical cure is shown in Figure 8. Mean rankings and 95% credibility intervals are shown as a forest plot (Fig. 9), and as a table in Supplementary File S4.

Fig. 8. (a–d) The distribution of the probability of clinical cure in the 5000 simulations in the network meta-analysis examining the relative efficacy of antimicrobial treatments given for clinical mastitis, shown for treatments available as a currently labeled intramammary product in North America.

Fig. 9. Forest plot of mean rank and 95% credibility interval for the network meta-analysis examining the relative efficacy (clinical cure) of antimicrobial treatment of clinical mastitis in lactating dairy cattle, shown for treatments available as a currently labeled intramammary product in North America. Full treatment arm descriptions are found in Table 1.
Table 5. Risk ratio comparison of all interventions assessed in the network meta-analysis for the outcome of clinical cure
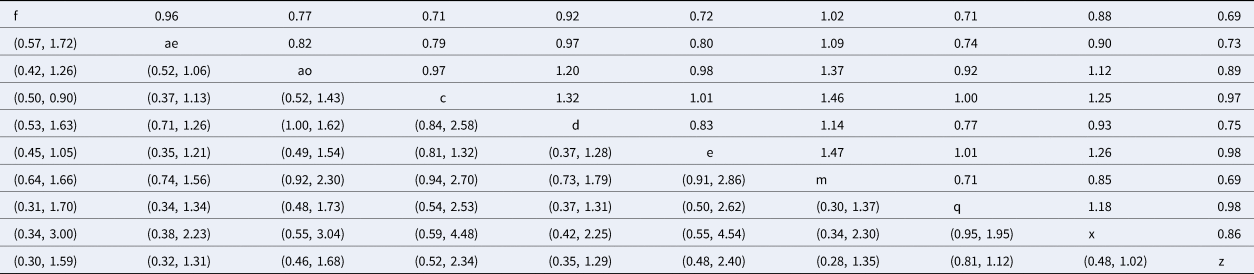
The upper right-hand section of the table represents the risk ratio between the numerator (upper left treatment) and denominator (lower right treatment). The lower left section of the table represents the 95% credibility interval for the comparison, with the rows and columns reversed. For example, the risk ratio for IMI at calving for treatment f (non-treated control) compared to treatment c (intramammary ceftiofur once daily for 2 days) is 0.71 (95% CI 0.50–0.90).
Discussion
Summary of evidence
As treatment of clinical mastitis reflects a large proportion of antimicrobial use in the dairy cattle industry, optimizing the use in this area can have a significant impact on overall Antimicrobial use (AMU). Evidence around efficacy can allow for the selection of an equally efficacious product of lesser importance to human health, and avoiding selection of ineffective antimicrobials can reduce unnecessary use where there is no benefit to animal health or welfare. Based on the information in the network, although most antimicrobial treatments were numerically superior to non-treated controls, large credibility intervals meant that no treatments were statistically different than each other or non-treated controls. As a result, we cannot conclude whether our results for this outcome were due to treatments being equivalent, or were due to a lack of power within the network. This could be further confounded by pathogen causing the clinical mastitis, given the small number of trials. This review provides information on the geometry of the networks, and identifies current gaps which can be addressed in future work in order to build the value of both future work and the entire body of evidence.
Limitations of the body of evidence
Although a large number of trials evaluated the outcomes of bacteriologic and clinical cure, and included the major species groups per-defined, the lack of replication of interventions lead to disparate networks in the case of bacteriologic cure, and contributed to the lack of precision in the clinical cure network. In order to utilize the information from all trials, each trial must connect to at least one other trial through at least one common intervention arm. Future research could consider the geometry of these networks, and if disconnected intervention arms are of importance, trials with one arm connecting to a larger body of evidence would serve to increase the value of the work conducted beyond the comparisons in a single trial. Replication of interventions would also increase the precision of point estimates which would allow for a better assessment of relative efficacy.
Within the clinical cure network, there was variation in both the eligibility criteria for cases among trials, and in the measurement of the outcome. These sources of variation both have the potential to contribute to heterogeneity in the analyses. Without adequate precision, it is not possible to assess heterogeneity within any pairwise comparison between two intervention arms in the network. Additionally, if it had been possible to analyze networks for different bacteriologic cure for one or more species, it would have been interesting to contrast the results with the results from the clinical cure network, as there was more (but not absolute) similarity between definition of cases and cure within each network for the bacterial cure outcomes.
Potential for bias arising from missing outcome data was seen in many trials for both outcomes, often due to a lack of reporting the number of study units analyzed. Although the information on randomization was not reported in a large number of trials, those which did report randomization often included the method used to generate the randomization sequence (i.e. few studies included the word ‘random’ without providing supporting evidence). Selective reporting of results was also challenging to assess, as without assessment of a trial protocol developed a priori, it is not possible to determine the potential for a multiplicity of outcome definitions or analyses. However, it is likely that the vast majority of, if not all, clinical trials in this area do have protocols developed prior to study conduct, in order to obtain ethical approval for use of animals. Including a time-stamped PDF documenting the original study protocol as supplementary material would aid the reader or reviewer in the interpretation of the work.
A lack of reporting key design features is consistent with recent evaluations of animal health research (Totton et al., Reference Totton, Cullen, Sargeant and O'Connor2018; Winder et al., Reference Winder, Churchill, Sargeant, LeBlanc, O'Connor and Renaud2019), and a lack of reporting has been associated with exaggerated treatment effects (Wellman and O'Connor, Reference Wellman and O'Connor2007; Sargeant et al., Reference Sargeant, Elgie, Valcour, Saint-Onge, Thompson, Marcynuk and Snedeker2009a, Reference Sargeant, Saint-Onge, Valcour, Thompson, Elgie, Snedeker and Marcynuk2009b). Not only do deficiencies in reporting preclude assessment for potential bias, but failure to report characteristics of data may also prevent inclusion in the meta-analysis. Indeed, in 7/54 (13%) of eligible trials, data were not able to be extracted for either bacteriologic or clinical cure. Reporting guidelines, such as the Reporting guidElines For randomized control trials in livEstoCk and food safTey (REFLECT) statement, have been designed in order to improve the quality of reporting of trials (O'Connor et al., Reference O'Connor, Sargeant, Gardner, Dickson, Torrence, Dewey, Dohoo, Evans, Gray, Greiner, Keefe, Lefebvre, Morley, Ramirez, Sischo, Smith, Snedeker, Sofos, Ward and Wills2010; Sargeant et al., Reference Sargeant, O'Connor, Gardner, Dickson and Torrence2010). Adherence to reporting guidelines will improve the utility of future research.
Limitations of the review
Only English-language studies were included, and therefore this review may not reflect the entirety of literature assessing the efficacy of lactating cow therapy on the bacteriologic and clinical cure of clinical mastitis. Additional studies would have increased the precision of our estimates. Decisions on what constituted unique treatments would influence potential heterogeneity within comparisons, but we attempted to be transparent on how decisions were made to group data. Challenge trials were not considered eligible for this review. While they provide proof of concept, we did not feel they could be combined with natural disease exposure studies to determine relative efficacy under field conditions (Sargeant et al., Reference Sargeant, Kelton and O'Connor2014a, Reference Sargeant, Kelton and O'Connor2014b), and may be more likely to show beneficial effects (Wisener et al., Reference Wisener, Sargeant, O'Connor, Faires and Glass-Kaastra2014).
Conclusions
From the network of evidence, there were no definitive conclusions regarding the relative efficacy of intramammary lactating cow therapy for clinical mastitis. This was primarily driven by a lack of precision of estimates for clinical cure and disparate networks for bacteriologic cures, meaning that information from several treatment comparisons could not contribute to a larger body of evidence of relative efficacy. Results from the clinical cure outcome network could be further confounded by pathogen causing the clinical mastitis, given the small number of trials. Consideration of the geometry of these networks can serve to inform future research, as both replication of important intervention arms and consideration of connection to existing networks would improve the future ability to determine relative efficacy. This would provide value beyond the comparisons within a given study, increasing the utility of both current and previous work. Challenges in the evaluation of bias in primary research stemmed from a lack of reporting. Consideration of reporting guidelines would also improve the utility of future research.
Supplementary material
The supplementary material for this article can be found at https://doi.org/10.1017/S1466252319000318.
Author contributions
CBW developed the review protocol, conducted relevance screening, extracted data, conducted risk of bias assessments, interpreted results, and wrote the manuscript drafts. JMS assisted with the development of the review protocol, conducted relevance screening, interpreted results, commented on manuscript drafts and approved the final manuscript. DH conducted the data analysis, provided guidance for interpretation of results, commented on manuscript drafts and approved the final manuscript. CW provided guidance on the conduct of the analysis and interpretation of results, and approved the final manuscript. KJC conducted relevance screening, extracted data, conducted risk of bias assessments, commented on manuscript drafts, and approved the final manuscript version. AMOC, DFK, and MAG assisted with the development of the review protocol, provided guidance on the interpretation of results, commented on manuscript drafts, and approved the final manuscript.
Financial support
This work was supported through a grant from the Ontario Ministry of Agriculture, Food, and Rural Affairs (OMAFRA) – University of Guelph Research Program (UofG2016-2643) and from support by the Dairy Farmers of Ontario.
Conflict of interest
None of the authors have conflicts to declare.