Introduction
Maternal psychological stress (or prenatal stress) during pregnancy, such as symptoms of anxiety and/or depression, has been associated with fetal programming of adverse long-term consequences for the child including an increased risk of fetal growth restriction (for example, Steer et al. Reference Steer, Scholl, Hediger and Fischer1992; Rondo et al. Reference Rondo, Ferreira, Nogueira, Ribeiro, Lobert and Artes2003; Khashan et al. Reference Khashan, McNamee, Abel, Pedersen, Webb, Kenny, Mortensen and Baker2008; Liu et al. Reference Liu, Murphy, Murtha, Fuemmeler, Schildkraut, Huang, Overcash, Kurtzberg, Jirtle, Iversen, Forman and Hoyo2012), emotional and behavioural problems, learning difficulties, cognitive impairment and psychopathology (for reviews, see Van den Bergh et al. Reference Van den Bergh, Mulder, Mennes and Glover2005; Talge et al. Reference Talge, Neal and Glover2007). In addition, there is evidence that prenatal depression can impair the normal development of postnatal maternal behaviour and mother–infant interactions (Pearson et al. Reference Pearson, Melotti, Heron, Joinson, Stein, Ramchandani and Evans2012).
Located at the boundary between the maternal and fetal environments, the placenta supports fetal growth and development through numerous functions including transport of nutrients and oxygen and the production and metabolism of hormones (Gude et al. Reference Gude, Roberts, Kalionis and King2004). The placenta has been proposed as a potential mechanism mediating the association between maternal prenatal stress and adverse infant outcomes (O'Donnell et al. Reference O'Donnell, O'Connor and Glover2009; Janssen et al. Reference Janssen, Kertes, McNamara, Braithwaite, Creeth, Glover and John2016). Consistent with this hypothesis, prenatal stress has previously been associated with altered placental function in both animal models (Mairesse et al. Reference Mairesse, Lesage, Breton, Breant, Hahn, Darnaudery, Dickson, Seckl, Blondeau, Vieau, Maccari and Viltart2007; Jensen Pena et al. Reference Jensen Pena, Monk and Champagne2012) and in humans (O'Donnell et al. Reference O'Donnell, Bugge Jensen, Freeman, Khalife, O'Connor and Glover2012; Blakeley et al. Reference Blakeley, Capron, Jensen, O'Donnell and Glover2013; Reynolds et al. Reference Reynolds, Pesonen, O'Reilly, Tuovinen, Lahti, Kajantie, Villa, Laivuori, Hamalainen, Seckl and Raikkonen2015).
Imprinted genes, which are monoallelically expressed with expression depending on the parent of origin (Surani, Reference Surani1998), are regulated by epigenetic marks that may respond to in utero environmental stimuli. These genes have been suggested as potential mediators of adverse infant outcomes because of their well-established roles in controlling fetal growth, placental development, adult behaviour and metabolism (Lefebvre et al. Reference Lefebvre, Viville, Barton, Ishino, Keverne and Surani1998; Li et al. Reference Li, Keverne, Aparicio, Ishino, Barton and Surani1999; Curley et al. Reference Curley, Pinnock, Dickson, Thresher, Miyoshi, Surani and Keverne2005; Smith et al. Reference Smith, Garfield and Ward2006; Tunster et al. Reference Tunster, Jensen and John2013; Jensen et al. Reference Jensen, Tunster and John2014; McNamara & Isles, Reference McNamara and Isles2014). Aberrant expression of imprinted genes in the placenta has also been demonstrated to be associated with impaired infant neurobehavioural development in humans (Green et al. Reference Green, Kappil, Lambertini, Armstrong, Guerin, Sharp, Lester, Chen and Marsit2015). Recent studies demonstrate that a subset of imprinted genes converge on the endocrine lineages of the mouse placenta to regulate placental hormone production (John, Reference John2013). This may be of particular relevance to maternal prenatal stress as the placenta is a significant source of hormones, such as placental lactogens (Glynn & Sandman, Reference Glynn and Sandman2011). These lactogenic hormones act on the maternal brain, priming the mother for pregnancy and postnatal care (Bridges et al. Reference Bridges, DiBiase, Loundes and Doherty1985, Reference Bridges, Numan, Ronsheim, Mann and Lupini1990, Reference Bridges, Robertson, Shiu, Sturgis, Henriquez and Mann1997; Bridges & Freemark, Reference Bridges and Freemark1995; Glynn & Sandman, Reference Glynn and Sandman2011). In addition, impaired placental lactogen production has been associated with adverse infant outcomes such as fetal growth restriction (Roh et al. Reference Roh, Budhraja, Kim, Nelson and Sadovsky2005; Dutton et al. Reference Dutton, Warrander, Roberts, Bernatavicius, Byrd, Gaze, Kroll, Jones, Sibley, Froen and Heazell2012). Thus, it is possible that aberrant placental imprinted gene expression and resulting impaired placental lactogen production mediate the association between maternal prenatal stress and adverse infant outcomes. In support of such a hypothesis, epigenetic changes in cord blood DNA at imprinted loci have been associated with both depressed maternal mood (Liu et al. Reference Liu, Murphy, Murtha, Fuemmeler, Schildkraut, Huang, Overcash, Kurtzberg, Jirtle, Iversen, Forman and Hoyo2012) and maternal stress (Vidal et al. Reference Vidal, Benjamin Neelon, Liu, Tuli, Fuemmeler, Hoyo, Murtha, Huang, Schildkraut, Overcash, Kurtzberg, Jirtle, Iversen and Murphy2014). The expression of imprinted genes in the placenta has not previously been examined in relation to maternal prenatal stress.
In this study we examined the expression levels of the paternally expressed gene 3 (PEG3), paternally expressed gene 10 (PEG10), pleckstrin homology-like domain family a member 2 (PHLDA2) and cyclin-dependent kinase inhibitor 1C (CDKN1C). These four imprinted genes were chosen based on the conserved imprinting status between mouse and human and their predicted role in regulating the production of placental hormones, including the placental lactogens, known to induce physiological changes in pregnant women (John, Reference John2013). Expression of these genes was first analysed in a pilot cohort of women (the Manchester cohort) with clinically diagnosed depression during pregnancy, with results suggesting a significant alteration in placental PEG3. Based on these results, we further analysed placental PEG3 expression in two additional independent cohorts of mothers reporting prenatal symptoms of depression. Finally, given the proposed link between PEG3 expression and placental hormones, we quantified expression of human placental lactogen (hPL, also known as chorionic somatomammotropins; CSH) in all three cohorts.
Method
Manchester cohort
Placental gene expression was first analysed in a pilot cohort. Women (n = 75) presenting with maternal perception of reduced fetal movements (RFM) at St Mary's Hospital (Manchester, UK) were approached to participate in the study as previously described (Dutton et al. Reference Dutton, Warrander, Roberts, Bernatavicius, Byrd, Gaze, Kroll, Jones, Sibley, Froen and Heazell2012; Warrander et al. Reference Warrander, Batra, Bernatavicius, Greenwood, Dutton, Jones, Sibley and Heazell2012). Written informed consent was obtained and the study approved by Oldham and Greater Manchester North Research Ethics Committees (REC no. 08/1011/83 and 11/NW/0664). A diagnosis of depression during pregnancy, including any treatment prescribed, was recorded from the participant's medical notes.
Queen Charlotte's and My Baby and Me study (MBAM) cohorts
For both cohorts, women awaiting an elective Caesarean section (with no known complications of pregnancy) were recruited from Queen Charlotte's maternity hospital, London. Participants in the Queen Charlotte's cohort (n = 40) were recruited between 2010 and 2011, while an independent cohort of women participating in the MBAM (n = 81) were recruited in 2014. Written informed consent was obtained. The Queen Charlotte's study was approved by the Ethics Committee of Hammersmith and Queen Charlotte's Hospital, London (REC no. 08/H0708/126) and MBAM was approved by the Research Ethics Committee of London (Chelsea) (REC no. 13/LO/1436).
At the time of recruitment, maternal prenatal depressive symptoms were measured using the Edinburgh Postnatal Depression Scale (EPDS). This questionnaire has been validated for use during pregnancy (Cox et al. Reference Cox, Chapman, Murray and Jones1996), with total EPDS scores ranging from 0 (low depression) to 30 (high depression). An EPDS score ⩾13 is used to identify women at risk of a depressive disorder (Cox et al. Reference Cox, Holden and Sagovsky1987).
Placental dissection
All placentas were collected immediately after delivery and sampled within 1 h (O'Donnell et al. Reference O'Donnell, Bugge Jensen, Freeman, Khalife, O'Connor and Glover2012; Warrander et al. Reference Warrander, Batra, Bernatavicius, Greenwood, Dutton, Jones, Sibley and Heazell2012). Villous trophoblast tissue samples were taken from the maternal surface of the placenta, midway between the cord and distal edge. Sampling methods were comparable across all three independent cohorts. Tissue samples were washed in phosphate-buffered saline to remove maternal blood and stored in RNAlater (Sigma-Aldrich, UK) prior to storage at −80°C.
Gene expression analysis
Total RNA was extracted from the placental tissue samples from the Manchester and MBAM cohorts using the GenElute Mammalian Total RNA Miniprep Kit (Sigma-Aldrich, UK). RNA quantity and quality (based on the 260:280 absorbance ratio) were assessed using a NanoDrop ND-1000 spectrophotometer. From the Queen Charlotte's cohort, RNA was extracted from placental tissue samples using RNeasy Mini Kits (Qiagen, UK). In addition, for this cohort RNA integrity was examined using a Bioanalyzer 2100 (Agilent, UK), according to the protocol for a RNA 6000 Nano Assay; samples with a RNA integrity number (RIN) value of ⩾5 were considered of sufficient quality for reverse transcription and quantitative polymerase chain reaction (qPCR).
For the Manchester cohort, 5 µg of RNA were reverse transcribed using M-MuLV reverse transcriptase (Promega, UK) with random hexamers, according to the manufacturer's instructions. For both the Queen Charlotte's and MBAM cohorts, 2 µg of RNA were reverse transcribed using the Superscript II first strand complementary DNA (cDNA) synthesis system (Invitrogen, UK), according to the manufacturer's instructions.
Quantitative RT-PCR was performed using a Chromo 4 Four Colour Real Time Detector (MJ Research) in a 20 µl reaction containing 5 µl of cDNA (diluted 1 in 50), 1X Buffer 2 mm MgCl2, 2 mm deoxynucleotides (dNTPs), 0.65 units Taq [Fermentas (Thermo), UK], 1 µ m of each primer (Sigma-Aldrich, UK) and 0.12X Sybr Green (Invitrogen, UK). All samples were run in triplicate and duplicate plates were performed for MBAM. Conditions for amplification were: (1) 15 min at 94°C; (2) 30 s at 94°C; (3) 30 s at 60°C; (4) 30 s at 72°C; and (5) 30 s 75°C, with steps 2–5 repeated for a total of 40 cycles. Melt Curve was performed from 70 to 94°C, reading every 0.5°C and holding for 2 s.
Primer sequences were as follows: YWHAZ forward: TTCTTGATCCCCAATGCTTC and reverse: AGTTAAGGGCCAGACCCAGT; PEG3 forward: CTCACAACACAATCCAGGAC and reverse: TAGACCTCGACTGGTGCTTG (Feng et al. Reference Feng, Marquez, Lu, Liu, Lu, Issa, Fishman, Yu and Bast2008); PEG10 forward: AAATTGCCTGACATGAAGAGGAGTCTA and reverse:AAGCCTAGTCACCACTTCAAAACACACTAAA (Diplas et al. Reference Diplas, Lambertini, Lee, Sperling, Lee, Wetmur and Chen2009); PHLDA2 forward: GAGCGCACGGGCAAGTA and reverse: CAGCGGAAGTCGATCTCCTT (Apostolidou et al. Reference Apostolidou, Abu-Amero, O'Donoghue, Frost, Olafsdottir, Chavele, Whittaker, Loughna, Stanier and Moore2007); CDKN1C forward: CCCATCTAGCTTGCAGTCTCTT and reverse: CAGACGGCTCAGGAACCATT (Diplas et al. Reference Diplas, Lambertini, Lee, Sperling, Lee, Wetmur and Chen2009); hPL forward: CATGACTCCCAGACCTCCTTC and reverse: TGCGGAGCAGCTCTAGATTG (Dutton et al. Reference Dutton, Warrander, Roberts, Bernatavicius, Byrd, Gaze, Kroll, Jones, Sibley, Froen and Heazell2012). hPL primers were designed to analyse the expression of CSH1/hPL-A and CSH2/hPL-B, from which the majority of circulating placental lactogens is derived (Newbern & Freemark, Reference Newbern and Freemark2011). Primer specificity was assessed based on gel electrophoresis product band size and qPCR melt curves.
Gene expression data are presented as the ∆CT (target gene expression relative to the housekeeping gene YWHAZ) and as the fold change in expression, calculated using the 2−∆∆CT (Livak & Schmittgen, Reference Livak and Schmittgen2001) where the ∆∆CT is the target gene expression relative to expression in the control group. The housekeeping gene YWHAZ has previously been demonstrated to be stably expressed in the human placenta (Meller et al. Reference Meller, Vadachkoria, Luthy and Williams2005; Murthi et al. Reference Murthi, Fitzpatrick, Borg, Donath, Brennecke and Kalionis2008; Cleal et al. Reference Cleal, Day, Hanson and Lewis2009, Reference Cleal, Day, Hanson and Lewis2010). Furthermore, we found no significant difference in YWHAZ expression between control and depressed participants in the Manchester cohort (p = 0.76) in an initial pilot study.
Data analysis
Placental gene expression and maternal EPDS data were normally distributed and parametric statistical tests were used to analyse the data. Placental gene expression data were not significantly associated with any maternal demographics listed in Table 1. The effect of potential confounders (infant birth weight, sex and gestational age) was examined using multiple linear regression analysis. To ease interpretation, ∆CT values representing gene expression values have been inverted [x(−1)] such that lower values represent decreasing gene expression. Mediation analysis was carried out in IBM SPSS Statistics Version 20 and PROCESS for SPSS version 2.15 (Hayes, Reference Hayes2013). Bootstrap confidence intervals (CIs) were used to assess statistical significance of the indirect effect (Hayes, Reference Hayes2013).
Table 1. Participant characteristics of the Manchester, Queen Charlotte's and MBAM cohorts
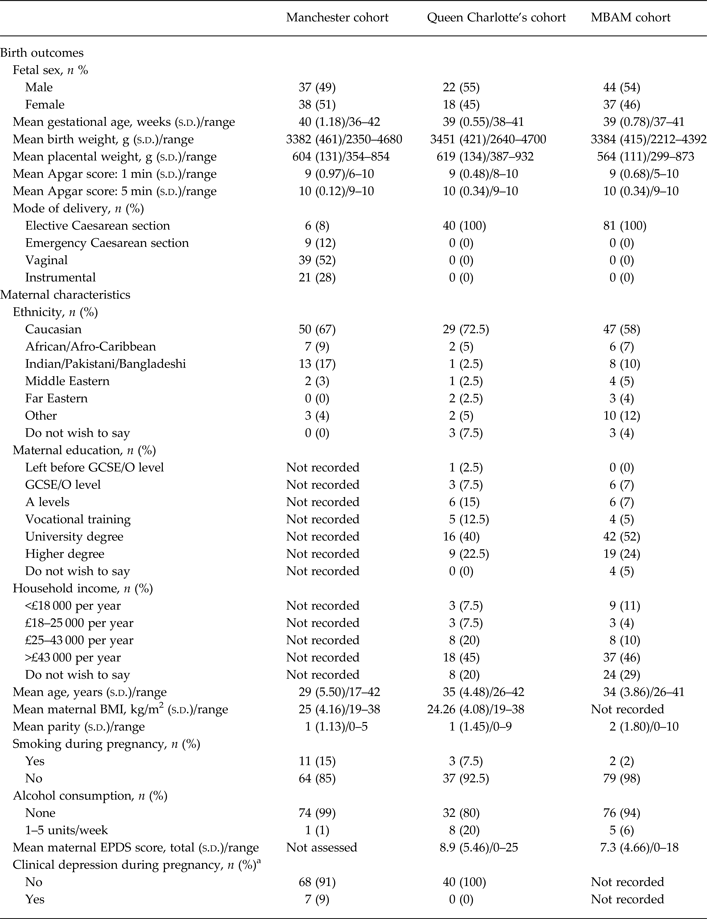
MBAM, My Baby and Me study; s.d., standard deviation; GCSE, General Certificate of Secondary Education; O level, Ordinary level; A level, Advanced level; BMI, body mass index; EPDS, Edinburgh Postnatal Depression Scale.
a A diagnosis of clinical depression was recorded from participants’ medical notes.
Results
Manchester cohort
In an initial pilot study, expression of the imprinted genes PEG3, PEG10, PHLDA2 and CDKN1C was analysed in a cohort of placentas (n = 75), which included a subset of women (n = 7) who had diagnosed depression during pregnancy. Characteristics of the study participants in the Manchester cohort are shown in Table 1. Participants with diagnosed depression did not differ significantly in terms of maternal demographics from control participants (results not shown).
There was no significant alteration in the expression of PEG10, PHLDA2 or CDKN1C in association with maternal depression (Fig. 1a ). However, placental PEG3 expression was significantly decreased (by 41%) in placentas from depressed participants compared with controls (Fig. 1a ). The association between placental PEG3 expression and maternal diagnosed depression remained significant after controlling for infant birth weight, offspring sex and gestational age (F 4,73 = 3.43, p = 0.01, R 2 = 0.16), with only maternal depression diagnosis significantly predicting placental PEG3 expression (p = 0.003). Three participants reported prescribed anti-depressant use during pregnancy; the observed decrease in placental PEG3 expression remained significant with the exclusion of these participants (p = 0.02, n = 75).

Fig. 1. Manchester cohort – maternal diagnosed depression and placental gene expression. (a) There was a significant decrease in paternally expressed gene 3 (PEG3) expression in women with diagnosed depression during pregnancy. Placental paternally expressed gene 10 (PEG10), pleckstrin homology-like domain family a member 2 (PHLDA2) and cyclin-dependent kinase inhibitor 1C (CDKN1C) expression was not significantly altered. (b) Human placental lactogen (hPL) expression was also significantly reduced in depressed participants. Values are means of fold gene expression, with standard errors represented by vertical bars. * p < 0.05.
Given the proposed regulation of placental lactogens by PEG3, expression of hPL was also analysed in this cohort. Placental PEG3 and hPL expression were significantly positively correlated (r = 0.26, p = 0.03, n = 75). There was a significant 44% decrease in placental hPL expression in depressed participants compared with controls (Fig. 1b ).
Queen Charlotte's cohort
Results from the pilot study indicated a significant decrease in placental PEG3 and hPL expression in a small number of women with clinically diagnosed depression during pregnancy. To determine whether more general symptoms of prenatal depression were similarly associated with aberrant placental gene expression of these two genes, we made use of a second independent cohort (the Queen Charlotte's cohort; n = 40) with information on maternal prenatal depression symptoms. The characteristics of the study participants in the Queen Charlotte's cohort are shown in Table 1. Indication for elective Caesarean section (ELCS) included previous section (n = 26, 65%), breech presentation (n = 7, 18%), maternal request (n = 2, 5%) and other (e.g. previous tear, low-lying placenta, n = 5, 12%). No participant reported prescribed anti-depressant use during pregnancy.
Despite promising pilot data, the modest correlation between placental PEG3 expression and maternal prenatal EPDS scores (r = −0.23, p = 0.16, n = 40) failed to reach statistical significance. Placental PEG3 expression does not differ between male and female placentas from normal, uncomplicated pregnancies (Janssen et al. Reference Janssen, Tunster, Savory, Holmes, Beasley, Parveen, Penketh and John2015) but male and female human fetuses are known to respond differently to an adverse intra-uterine environment (Clifton, Reference Clifton2010). Therefore, the association between placental PEG3 expression and maternal depression was also analysed independently in male and female pregnancies. When the PEG3 expression data were analysed according to fetal sex, this revealed a significant negative association between placental PEG3 expression and maternal EPDS scores in males (p = 0.048) but not females (p = 0.74) (Fig. 2a and b ). A 13% decrease in PEG3 expression was observed in placentas from participants with an EPDS score ⩾13, highlighting them as at risk of a depressive disorder, although this difference did not reach statistical significance (p = 0.16) possibly due to the relatively small number of women scoring above this cut-off (n = 9). However, when participants in the current study were grouped into lowest (mean EPDS 2.7, n = 15) and highest (mean EPDS 14.5, n = 15) EPDS scorers there was a significant 15% decrease (p = 0.03) in PEG3 expression in placentas from the highest EPDS scorers compared with the lowest EPDS scorers (Fig. 2c ).

Fig. 2. Queen Charlotte's cohort – maternal depression symptoms and placental gene expression. The association between maternal depression and placental paternally expressed gene 3 (PEG3) expression appears to be sex specific with a significant inverse association between Edinburgh Postnatal Depression Scale (EPDS) scores and PEG3 expression in male (a) but not female (b) placentas. There was a significant decrease in PEG3 (c) and human placental lactogen (hPL) (d) expression between 15 participants with the lowest and highest EPDS scores. Values are means of fold gene expression, with standard errors represented by vertical bars. * p < 0.05.
Placental hPL expression was also negatively correlated with maternal EPDS scores (r = −0.27, p = 0.13, n = 40) but without statistical significance in the full cohort or when analysed independently in male (r = −0.25, p = 0.25, n = 22) and female (r = −0.29, p = 0.26, n = 17) placentas. However, as with PEG3, a significant 31% decrease (p = 0.03) in placental hPL expression was observed in the highest EPDS scorers compared with the lowest EPDS scorers (Fig. 2d ). There was no significant correlation between placental PEG3 and hPL expression (r = −0.13, p = 0.43, n = 40).
MBAM cohort
We next sought to replicate these findings in the MBAM cohort (n = 81), a larger independent cohort of samples with similar information on maternal prenatal depression symptoms. Characteristics of the study participants in the MBAM cohort are shown in Table 1. Indication for ELCS included previous Caesarean section (n = 43, 53%), breech presentation (n = 15, 19%), maternal request (n = 11, 14%), placenta previa (n = 6, 7%) and obstetric history (n = 6, 7%).
Placental PEG3 expression was significantly inversely associated with maternal prenatal EPDS scores (r = −0.32, p = 0.003, n = 81). As with the Queen Charlotte's cohort, when gene expression was analysed according to fetal sex, there was a significant inverse association between PEG3 expression and maternal EPDS scores in male (r = −0.42, p = 0.005, n = 44) but not female placentas (r = −0.22, p = 0.19, n = 37) (Fig. 3a and b ). Multiple linear regression analysis showed that the association between placental PEG3 expression and maternal depression symptoms remained significant after controlling for infant birth weight, offspring sex and gestational age (F 4,76 = 2.45, p = 0.05, R 2 = 0.11), with only maternal prenatal EPDS scores significantly predicting placental PEG3 expression (p = 0.005). Only two participants reported prescribed anti-depressant use during pregnancy; the association between placental PEG3 expression and maternal EPDS scores remained significant with the exclusion of these participants (r = −0.30, p = 0.006, n = 79). Finally, a significant 22% decrease in PEG3 expression was observed in placentas from participants with EPDS scores ⩾13, highlighting them as at risk of a depressive disorder (Fig. 3c ).

Fig. 3. My Baby and Me Study (MBAM) cohort – maternal depression symptoms and placental gene expression. The inverse association between paternally expressed gene 3 (PEG3) expression and maternal depression was significant in male (a) but not female (b) placentas. There was a significant decrease in PEG3 (c) and human placental lactogen (hPL) (d) expression in participants with Edinburgh Postnatal Depression Scale (EPDS) scores ⩾13 (cut-off for clinical depression). Values are means of fold gene expression, with standard errors represented by vertical bars. * p < 0.05.
Placental hPL expression was also significantly inversely associated with maternal prenatal EPDS scores (r = −0.36, p = 0.001, n = 81) in the overall cohort. As with PEG3, this association was sex specific, being significant in male (r = −0.51, p < 0.001, n = 44) but not female (r = −0.11, p = 0.53, n = 37) placentas. There was also a significant 21% decrease in hPL expression in placentas from participants with EPDS scores ⩾13 (Fig. 3d ). Finally, there was a positive correlation between placental PEG3 and hPL expression although this was not statistically significant in the overall cohort (r = 0.20, p = 0.08, n = 81), being significant in male (r = 0.39, p = 0.01, n = 44) but not female placentas (r = −0.26, p = 0.12, n = 37).
Mediation analysis
Mediation analysis determines the effect of an independent variable on a dependent variable via a third mediating variable and is typically conducted in larger cohort sizes of >100 participants (Fritz & Mackinnon, Reference Fritz and Mackinnon2007). In this study mediation analysis was carried out in the two largest cohorts (Manchester and MBAM) with the hypothesis that the association between placental PEG3 and maternal EPDS scores is mediated by placental hPL expression. In the Manchester cohort (n = 75) the indirect effect of placental PEG3 expression on maternal EPDS scores mediated by placental hPL expression was not statistically significant in the overall cohort (B = 0.29, 95% CI −0.08 to 1.13) or when only male placentas were analysed (B = 0.91, 95% CI −0.64 to 5.20). Similarly, in the MBAM cohort (n = 81) the indirect effect of placental PEG3 expression was not statistically significant in the overall cohort (B = −0.53, 95% CI −1.71 to 0.46) or when only male placentas were analysed (B = 1.35, 95% CI −0.26 to 3.64).
Maternal depression and birth weight
Maternal stress is known to be associated with an increased risk of fetal growth restriction (Steer et al. Reference Steer, Scholl, Hediger and Fischer1992; Rondo et al. Reference Rondo, Ferreira, Nogueira, Ribeiro, Lobert and Artes2003; Khashan et al. Reference Khashan, McNamee, Abel, Pedersen, Webb, Kenny, Mortensen and Baker2008; Liu et al. Reference Liu, Murphy, Murtha, Fuemmeler, Schildkraut, Huang, Overcash, Kurtzberg, Jirtle, Iversen, Forman and Hoyo2012) and therefore we analysed birth weight in relation to maternal depression in the three cohorts. Birth weight was not significantly altered in women with diagnosed depression (3.30 kg v. 3.09 kg, p = 0.41, n = 75) in the Manchester cohort. There was also no significant association between maternal prenatal EPDS scores and birth weight in the Queen Charlotte's (r = −0.17, p = 0.28, n = 40) or MBAM (r = 0.12, p = 0.28, n = 81) cohorts. Reduced expression of Peg3 in mice has also been linked to fetal growth restriction (Li et al. Reference Li, Keverne, Aparicio, Ishino, Barton and Surani1999). There was no significant association between placental PEG3 expression and birth weight in the Manchester (r = 0.05, p = 0.70, n = 75), Queen Charlotte's (r = 0.09, p = 0.58, n = 40) or MBAM (r = −0.02, p = 0.84, n = 81) cohorts. However, it should be noted that for the Queen Charlotte's and MBAM cohorts women with complications of pregnancy (including fetal growth restriction) were excluded and that previous studies demonstrating an association between maternal prenatal stress and fetal growth restriction examined substantially larger cohorts (Talge et al. Reference Talge, Neal and Glover2007).
Discussion
This study has identified a novel association between maternal prenatal depression and decreased placental expression of the imprinted gene PEG3. In our pilot study, placental expression of the imprinted genes PEG3, PEG10, PHLDA2 and CDKN1C was examined in relation to clinically diagnosed depression. Despite the small number of participants, there was a clear significant reduction in placental PEG3 expression but no alteration in expression of the other three imprinted genes examined. Importantly, placental PEG3 expression was also significantly inversely associated with symptoms of maternal prenatal depression in two further independent cohorts, the Queen Charlotte's and MBAM cohorts. This is the first report linking abnormal expression of this imprinted gene in the placenta with maternal depression during pregnancy, which may inform our understanding of the mechanisms underlying the association between prenatal depression and adverse offspring outcomes.
PEG3 exemplifies all the known functions of imprinted genes; it acts to regulate fetal growth, placental development, behaviour and metabolism in mice (Li et al. Reference Li, Keverne, Aparicio, Ishino, Barton and Surani1999; Curley et al. Reference Curley, Pinnock, Dickson, Thresher, Miyoshi, Surani and Keverne2005; Champagne et al. Reference Champagne, Curley, Swaney, Hasen and Keverne2009; Chiavegatto et al. Reference Chiavegatto, Sauce, Ambar, Cheverud and Peripato2012; Kim et al. Reference Kim, Frey, He, Kim, Ekram, Bakshi, Faisal, Perera, Ye and Teruyama2013). We did not observe reduced birth weight in this study, probably as a result of the small study numbers and exclusion of growth-restricted infants. However, it is possible that reduced expression of PEG3 in human pregnancy could contribute to fetal growth restriction, either directly or indirectly via a placental defect, and therefore mediate the previously reported association between maternal prenatal stress and impaired fetal growth.
In this study we also demonstrated a significant inverse association between placental hPL expression and maternal prenatal depression in all three cohorts. hPL is a lactogenic hormone, produced in large quantities by the placenta, and is important for maternal glucose management during pregnancy and fetal growth (Newbern & Freemark, Reference Newbern and Freemark2011). Maternal serum hPL levels and placental hPL expression have previously been demonstrated to be significantly reduced in pregnancies complicated by fetal growth restriction (Roh et al. Reference Roh, Budhraja, Kim, Nelson and Sadovsky2005; Dutton et al. Reference Dutton, Warrander, Roberts, Bernatavicius, Byrd, Gaze, Kroll, Jones, Sibley, Froen and Heazell2012). Placental lactogens are closely related to pituitary prolactin (Haig, Reference Haig2008). This hormone family is known to be important for driving the maternal physiological adaptations to pregnancy (John, Reference John2013) but studies in rodents suggest that these hormones may also be important for psychological adaptation to pregnancy. Both prolactin and placental lactogen can induce maternal behaviour in non-pregnant rodents (Bridges et al. Reference Bridges, DiBiase, Loundes and Doherty1985, Reference Bridges, Numan, Ronsheim, Mann and Lupini1990, Reference Bridges, Robertson, Shiu, Sturgis, Henriquez and Mann1997; Bridges & Freemark, Reference Bridges and Freemark1995) and prolactin plays an important role in pregnancy-related neurogenesis (Bridges & Grattan, Reference Bridges and Grattan2003; Shingo et al. Reference Shingo, Gregg, Enwere, Fujikawa, Hassam, Geary, Cross and Weiss2003; Walker et al. Reference Walker, Vukovic, Koudijs, Blackmore, Mackay, Sykes, Overall, Hamlin and Bartlett2012). There has not yet been a comprehensive examination of serum hPL levels in relation to maternal mood in humans. However, decreased maternal serum levels of prolactin have been reported in human mothers with postnatal depression symptoms (Abou-Saleh et al. Reference Abou-Saleh, Ghubash, Karim, Krymski and Bhai1998; Ingram et al. Reference Ingram, Greenwood and Woolridge2003; Groer & Morgan, Reference Groer and Morgan2007) and increased levels in mothers with low anxiety scores during pregnancy (Asher et al. Reference Asher, Kaplan, Modai, Neri, Valevski and Weizman1995). Prolactin contributes to a suppression of anxiety-related behaviours during pregnancy via binding to prolactin receptors, which are also known to bind the placental lactogens (Torner et al. Reference Torner, Toschi, Pohlinger, Landgraf and Neumann2001). As with reduced expression of PEG3, reduced placental lactogen production could contribute to a suboptimal pregnancy.
PEG3 has been proposed to regulate the production of placental hormones (John, Reference John2013). We observed a significant correlation between placental PEG3 and hPL expression in the two largest cohorts (Manchester and MBAM). Mediation analysis was carried out in these two cohorts with the hypothesis that the association between placental PEG3 and maternal depression is mediated by placental hPL expression. The indirect effect of placental PEG3 expression on maternal depression mediated by placental hPL expression was not statistically significant in either cohort. However, it should be noted that the sample sizes used in this analysis (Manchester cohort n = 75 and MBAM n = 81) are relatively low given that mediation analysis is typically conducted in studies of >100 participants (Fritz & Mackinnon, Reference Fritz and Mackinnon2007). While it is possible that the gene expression changes observed are independently related to maternal prenatal depression, studies in the mouse placenta demonstrate reduced expression of Peg3 drives reduced expression of Prls (mouse placental lactogens) (Broad & Keverne, Reference Broad and Keverne2011; Kim et al. Reference Kim, Frey, He, Kim, Ekram, Bakshi, Faisal, Perera, Ye and Teruyama2013). Future studies employing mediation analysis will probably be more informative in larger cohorts and will be crucial in determining the relationship between PEG3 and hPL in the human placenta.
The association between maternal prenatal depression symptoms and placental PEG3 and hPL expression showed a sex bias for both genes, in both cohorts, with the effect more marked with male fetuses. This result is of interest as previous studies suggest sexual dimorphism in infant outcomes following maternal prenatal stress (for a review, see Glover & Hill, Reference Glover and Hill2012). Boys, but not girls, are at an increased risk of attention-deficit/hyperactivity disorder (Li et al. Reference Li, Olsen, Vestergaard and Obel2010), schizophrenia (van Os & Selten, Reference van Os and Selten1998) and impaired motor development (Gerardin et al. Reference Gerardin, Wendland, Bodeau, Galin, Bialobos, Tordjman, Mazet, Darbois, Nizard, Dommergues and Cohen2011). There is also some preliminary evidence to suggest that women who give birth to boys are more likely to suffer from postpartum depression than those having girls (de Tychey et al. Reference de Tychey, Briancon, Lighezzolo, Spitz, Kabuth, de Luigi, Messembourg, Girvan, Rosati, Thockler and Vincent2008; Lagerberg & Magnusson, Reference Lagerberg and Magnusson2012). It will therefore be important to establish whether there is an association between placental PEG3 and hPL expression, fetal sex, infant outcomes and maternal postnatal depression.
This study was not designed to establish cause-or-effect relationships. Prenatal depression, or other factors associated with prenatal depression, may drive reduced expression of placental PEG3 and hPL in the placenta (Fig. 4: model 1). Peg3 in the rodent placenta is known to be responsive to environmental stimuli, such as maternal diet (Broad & Keverne, Reference Broad and Keverne2011; Radford et al. Reference Radford, Isganaitis, Jimenez-Chillaron, Schroeder, Molla, Andrews, Didier, Charalambous, McEwen, Marazzi, Sassoon, Patti and Ferguson-Smith2012). Alternatively, reduced PEG3 may contribute to or even initiate prenatal depression (Fig. 4: model 2). Loss of function of Peg3 in mice results in abnormal maternal behaviour (Li et al. Reference Li, Keverne, Aparicio, Ishino, Barton and Surani1999; Champagne et al. Reference Champagne, Curley, Swaney, Hasen and Keverne2009; Chiavegatto et al. Reference Chiavegatto, Sauce, Ambar, Cheverud and Peripato2012). However, in this scenario it is the dams that are mutant for Peg3 and not the placenta. Rather, aberrant placental PEG3 expression may contribute to abnormal maternal mood in human pregnancies, via impaired placental production of hPL (Janssen et al. Reference Janssen, Kertes, McNamara, Braithwaite, Creeth, Glover and John2016). It is also possible that both models are correct. A suboptimal maternal environment, which may include maternal prenatal stress, could misprogramme placental expression of PEG3, which in turn may alter placental signalling via hPL, thereby further contributing to the depressed mood (Fig. 4: model 3).

Fig. 4. Paternally expressed gene 3 (PEG3), human placental lactogen (hPL) signalling and maternal psychological adaptation to pregnancy. Model 1: prenatal depression causes reduced PEG3 and reduced hPL expression. Model 2: reduced PEG3 initiates prenatal depression through regulation of placental lactogen production. Model 3: an adverse intra-uterine environment causes reduced PEG3 expression limiting placental signalling and establishing a cycle of aberrant placental gene expression, aberrant signalling and abnormal maternal mood.
There are limitations to this study, primarily in the number of participants. However, this is countered by the reproducibility of these findings in three independent studies. Also, in the Queen Charlotte's and MBAM cohorts, maternal prenatal depression symptoms were assessed 1 day prior to elective Caesarean section when mothers may be particularly anxious. Ideally, future studies would involve assessment of maternal mood at different time points throughout pregnancy in a larger number of participants to provide a better indication of maternal prenatal stress during pregnancy. Another study limitation is that participants in the Manchester cohort suffered from RFM before delivery and so are not representative of uncomplicated pregnancies. However, both controls and women with depression during pregnancy experienced RFM and therefore this was not causative of the differences in gene expression observed. Finally, as depression diagnosis in the Manchester cohort was based solely on a diagnosis reported within clinical records, we cannot exclude the presence of depression symptoms in the control cases. Future studies with measures of depression for all participants (as with the Queen Charlotte's and MBAM cohorts) as well as more detailed information on the nature of depression and medication during pregnancy will be important in confirming the association between aberrant placental gene expression and diagnosed depression.
Conclusions
In summary, this study is the first to report reduced expression of both PEG3 and hPL in the human placenta in relation to adverse maternal mood. Substantial indirect data from rodent models suggest that this relationship is pathologically relevant and may indeed be causal. Animal studies will be important in further establishing cause-and-effect relationships. Meanwhile, it will be important to validate our findings in a larger study cohort and, importantly, to examine the outcomes of pregnancies with reduced PEG3 expression, both for the child and for the mother. Measuring maternal serum levels of hPL will be instrumental in determining whether reduced placental hPL expression in the term placenta reflects reduced hormone serum level during pregnancy. This is of clinical relevance since it may be possible to use maternal serum hPL levels as a biomarker in combination with self-report questionnaires to identify mothers at high risk of maternal depression. Finally, our current findings are of broader interest as reduced expression of PEG3 could provide a mechanistic explanation for the co-occurrence of maternal depression, low birth weight and poorer outcomes for the offspring, a finding that will lead to a greater understanding of both the causes and consequences of prenatal depression.
Acknowledgements
The authors would like to acknowledge all the women who participated in this study and the midwives at St Mary's Hospital and Queen Charlotte's Hospital for their help with sample collection. The Manchester cohort was supported by Manchester National Institute for Health Research (NIHR) Biomedical Research. The Queen Charlotte's cohort was supported by the Medical Research Council (MRC) (Eurostress), National Institutes of Health (R01MH073842) and the Genesis Research Trust. The MBAM cohort was supported by the Genesis Research Trust. A.B.J. was supported by a Biotechnology and Biological Sciences Research Council (BBSRC) Doctoral Training Grants (DTG) studentship and subsequently MRC project grant MR/M013960/1. S.J.T. was supported by BBSRC project grant BB/J015156/1. L.E.C. was supported by an Imperial College London Ph.D. studentship and both L.E.C. and P.G.R were supported by the NIHR Imperial Biomedical Research Centre.
Declaration of Interest
None.







